
Research of the potential biomarkers in vaginal microbiome for persistent high-risk human papillomavirus infection
Introduction
Over 80% of sexually active women have been infected by one or more human papillomavirus (HPV) types at some point in their lives (1). Over 50% of HPV infections resolve within 6–12 months and 80% are resolved within 2–5 years (2), 10–20% of infections persist latently (3), and only 0.3–1.2% of the initial infections will eventually progress to invasive cervical cancer. The average time interval between the infection with a carcinogenic type of HPV and the development of cervical cancer is 25 to 30 years (4). Persistent infection with high-risk HPV (HR-HPV) is necessary but not sufficient for the development of cervical cancer (5). Additional factors correlated with HR-HPV persistence include immunodeficiency caused by HIV, smoking, taking oral contraceptives and, more recently reported, vaginal dysbiosis (6).
The relationship between chronic prostatitis/chronic pelvic pain syndrome and uterine microbiota, gut microbiome and prostatic secretion microbiome has been widely investigated. Although less is understood about the role of the vaginal microbiome (VM) in human disease, the human microbiome project has extensively examined it. The classically-defined normal VM is dominated by one or more Lactobacillus sp. However, in a state of dysbiosis, there is a marked reduction of Lactobacillus and a boosted diversity of bacteria, with an increased abundance of anaerobic bacterial species (7-9). The human vaginal ecosystem is a dynamic environment in which microbes can affect the host’s physiology, while the host’s physiology can also affect the composition and function of the VM (10). What’s more, it is reported that vaginal dysbiosis was associated with an increased risk of incident HPV infection, HPV persistence, and high-grade lesions and cancer (11). Thus, the search for a bacterial cause in HPV infection continued with the advent of 16S rRNA sequencing to molecularly identify bacteria (12).
HR-HPV screening is highly sensitive for detection of cervical intraepithelial neoplasia (CIN) 2+, but holds a limited specificity, resulting in a high risk of invasive examination and overtreatment. This is especially the case for young women because of a high rate of transient infections in them. To mitigate this, we wanted to determine the possible VM predictors and determines of viral persistence and clearance by investigating the VM in persistent HR-HPV infection, incident HR-HPV infection, uninfected women using the Illumina sequencing platform. Besides, we want to explore the important role of the potential vaginal microbiota biomarker in the stratification algorithm for cervical cancer screening. Improved knowledge on these could lay the foundation for the novel diagnosis predictors and the avoiding of unnecessary invasive examinations, as well as the development of probiotics that may constitute a simpler and better prevention and/or restorative approach.
Methods
Ethics
Ethical approval was obtained from the Ethics Committee of Peking Union Medical College Hospital (PUMCH), Beijing, China (No. JS-1634). All experiments were performed in accordance with relevant guidelines and regulations. The registration No. in clinicaltrials.gov is NCT03548740. Written informed consent was obtained from all participants.
Study design
This prospective cohort study was implemented in a tertiary teaching hospital. According to the results of HPV test, the participants were divided into three groups. Group A: 59 cases with cervical persistent HPV infection (all the participants being persistently infected with the same kind of HR-HPV for at least 1 year until the time of sampling, and they haven’t been treated with antiviral drugs like α2b-recombinant human interferon, physiotherapy like laser therapy and cryotherapy, or surgical treatment like loop electrosurgical excision and cold knife conization). Group B: 139 cases with incident HPV infection (all the participants’ initial detection of HR-HPV was positive; or the participants had evidence of negative HPV infection during last year while the current test revealed positive HPV infection). Group C: 131 cases without HPV infection (the participants came to visit just for routine physical examination, and the test result of the current HPV status was negative).
Study population
The participants engaged in this research were those who visited the Department of Obstetrics & Gynecology of PUMCH between July 2018 and March 2019. All of the participants enrolled are women presenting for cervical cancer screening. Inclusion criteria: those aged 20 to 69 years old, having had vaginal intercourse for more than 3 years, and aren’t in menstrual, pregnancy or puerperium period. Exclusion criteria: Those who are virgin, having had total or subtotal hysterectomy, as well as the patients who were diagnosed with acute genital tract inflammation. Women who are HIV positive, have autoimmune disorders, or have a history of malignant tumors are also excluded. At the same time, all the participants should meet the following requirements: no vagina douching within the last 2 days, no vaginal intercourse within the last 3 days, no systemic application of antifungal agents or antibiotics or pessaries within the last 14 days of sampling.
Specimen collection
A sterile, disposable speculum was inserted without lubricant, and a sterile swab sample was taken from the posterior vaginal fornix and stored immediately under −80 °C for DNA extraction. At the same time, each patient was given a liquid Pap test with ThinPrep® Pap testing (Hologic, Inc., MA) and DNA capture via the Cobas® 4800 System HPV Genotyping Test (Roche Molecular Diagnostics, CA, USA) which is based on real-time qualitative PCR (RQ-PCR).
DNA extraction and the amplification of bacterial 16S rRNA V4 gene region and Illumina sequence were shown in Supplementary file 1.
Data analysis
SPSS 23.0 software (SPSS Inc., Chicago, IL, USA) was applied for the statistical analysis of the clinical data. Continuous variables were analyzed with t-test, and categorical variables were analyzed with Chi-square test. P<0.05 was interpreted to be statistically significant.
Results
Sociodemographic and clinical baseline characteristics
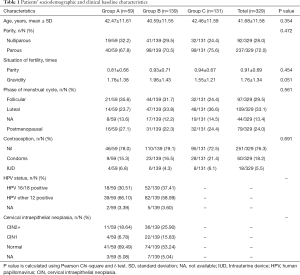
Baseline characteristics were generally similar within three groups. The mean ages of the three groups were 42.47±11.61, 40.59±11.55, and 42.46±11.59 years old respectively. There was no significant difference within the three groups regarding age (P=0.354), parity (P=0.454), gravidity (P=0.051), phase of menstrual cycle (P=0.561) and the method of contraception (P=0.691). Referring to the HPV infection status, there were 18 cases (30.51%) in group A and 52 cases (37.41%) in group B who are diagnosed with HPV 16/18 infection. Thirty-nine cases (66.10%) in group A and 82 cases (58.99%) in group B were diagnosed with other 12 types of HR-HPV infection (Table 1).
Full table
Microbiome community diversity
Identification of VM
A total of 65 phyla, 1,283 genera and 1,226 species were detected. In phylum level, the relative abundance of Actinobacteria, Proteobacteria, Bacteroidetes, and Tenericutes were higher in group A than group B and C. However, the relative abundance of Firmicutes was lower in group A than the other two groups (Figure 1A).
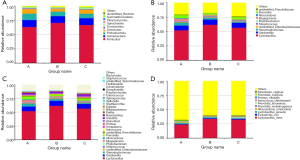
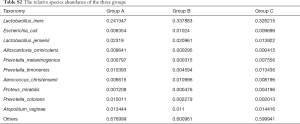
Lactobacillus was still the dominant genus in three groups (Figure 1B,C). In the top ten genera, group A has higher relative abundance than the other two groups in Stenotrophomonas, Megasphaera, Alloscardovia, and Prevotella, has lower relative abundance in Lactobacillus, Gardnerella, unidentified_Enterobacteriaceae, and Streptococcus (Table S1). To be emphasized, the abundance of Stenotrophomonas and Megasphaera were the highest in group A, followed by group B, while group C had the lowest abundance. Among the top ten genera, the abundance of Lactobacillus iners (L. iners) and Escherichia coli were lower in group A than in group B and C. The abundance of Lactobacillus jensenii, Alloscardovia omnicolens and Prevotella colorans were higher in group A than group B and C. It should be noted that, the abundance of Lactobacillus jensenii and Prevotella colorans were the highest in group A, followed by group B and the abundance in group C were the lowest. In reverse, group C had higher abundance than the other two groups in Prevotella melaninogenica, Prevotella timonensis, Proteus mirabilis, and Atopobium vaginae (Figure 1D, Table S2).

Full table
Full table
The structure of the VM within the three groups
Figure 2A showed that the species diversity increased along with the increasing of the sample size, and suggested that the sample size was adequate for analysis. From the rarefaction curve (Figure 2B), we can see that those from group C had the highest microbiome diversity, followed by group B, and group A had the lowest diversity. However, there was no significant difference within the three groups regarding the Shannon index (A vs. B, P=0.6161; A vs. C, P=0.4076; B vs. C, P=0.0886) (Figure 2C). The observed species are 490, 507 and 602 respectively in group A, B and C.

Identification of VM composition within the three groups
PCA (PC1 vs. PC2) revealed tighter clustering of group B and C’s microbiome compared to broad and variable clustering of microbiomes from group A (Figure 3A). Distance Matrix Heatmap based on the weighted UniFrac distance revealed that the VM’s difference between group A and B was the greatest, and that between group B and C was the smallest (Figure 3B). ANOSIM also validated this finding. There were significant differences between group A and B (R=0.06697, P=0.016), group A and C (R=0.05617, P=0.04). However, the difference between group B and C appeared to be less significant (R=0, P=0.415) (Figure 3C).
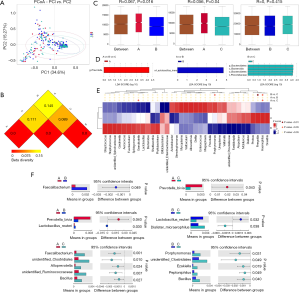
In the top 30 genera, the relative abundance of Phyllobacterium was significantly lower in group A than group B, but higher in group B than group C. Bacteroides had the lowest abundance in group A than B and C. Besides, the relative abundance of Prevotella, Porphyromonas and Enterococcus were significantly the highest in group A (Figures 3D,E,F,4, Table 2). Prevotella, Porphyromonas and Enterococcus are of highest relative abundance in group A, while Phyllobacterium and Bacteroides are of lowest abundance in group A (Figure 4, Table 2). In species level, a total of 32 species were found significantly different between group A and B, 4 of which had relative abundance above 0.1%. Forty-eight species were found significantly different between group A and C, and four had relative abundance above 0.1%. Forty-one species showed significant difference between group B and C, 4 of which had relative abundance above 0.1% (Figure 4). When analyzing the three groups altogether, Prevotella bivia (P. bivia), Enterococcus durans and Porphyromonas uenonis are significantly of highest relative abundance than the other two groups, while L. iners and Prevotella disiens (P. disiens) are significantly of lowest relative abundance than the other two groups (Figure 4, Table 2).
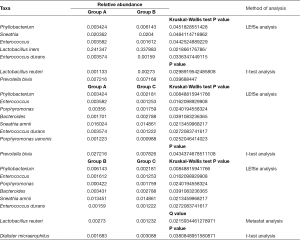
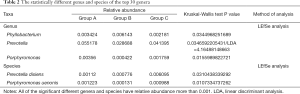
Full table
The diagnostic efficacy of the VM
There are 27 patients diagnosed with the other 12 types of HR-HPV persistent infection but normal ThinPrep cytology test (TCT) result, and three cases were finally confirmed with CIN2+. Thus, the diagnostic sensitivity is 100% while the specificity is only 11.11%. With the significantly different VM, we performed the receiver operating characteristic (ROC) curve to find the cut-off relative abundance of potential microbiome biomarker to help improve the diagnostic specificity. The specificity can reach to 66.67% and the sensitivity is still 100% when the relative abundance of P. bivia is over 0.05554% with P. disiens being under 0.02196%.
Discussion
It is unknown why some women develop persistent cervical HPV infections, but it is these women who are at the greatest risk of developing invasive cervical lesions. Cohort studies fail to identify individuals who resolve infection in a few days to a few weeks because of long sampling intervals (4–6 months). Data suggest that long-term persistence of HR-HPV may not always result in high-grade CIN (13). In the research for improved risk stratification of HPV-infected women who will ultimately develop cervical disease, it is worth considering whether identifying and characterizing any subset of super-responders would shed light on HPV persistence.
It has been shown that the reduction of genus Lactobacillus combined with increased diversity of VM has relation with HPV acquisition and persistence, as well as development of CIN and cervical cancer (6). It is those with the highest diversity of VM having the greatest instability (i.e., transition from one state to another) (14). However, there was also study revealing that neither variation in community composition, nor constantly high levels of apparent diversity (co-dominance) are necessarily indicative of dysbiosis (14). From our results, women with persistent HPV infection had the lowest diversity while uninfected women had the highest VM diversity, but there was no significant difference.
In the top 30 genera, persistent infection group had a lower relative abundance of Lactobacillus than the other two groups but without statistical difference. Five types of bacteria showed significant difference regarding their relative abundance within the three groups, including the obligated anaerobic bacteria Prevotella and Porphyromonas, facultative anaerobic bacteria Enterococcus, and aerobic bacteria Phyllobacterium. Genus Prevotella have been found in a variety of anatomic sites (15), and it was considered as a cause of infections. The genus Porphyromonas comprises several species of gram-negative anaerobes regarded as normal flora of the oral cavity and the gastrointestinal and genital tracts (16). The pathogenic potential of this genus varies among species.
Besides the findings in genera level, this study also revealed results of great importance in species level. A Lactobacillus-dominated VM protects women from adverse reproductive health outcomes. However, not all Lactobacillus are necessarily stable or “healthy”. L. iners is present in all women including those with “dysbiosis”, its beneficial role has therefore been debated (9,17,18). For example, in one study, a predominance of L. iners is a predicting factor for the development of bacterial vaginosis (BV) (19,20). L. iners predispose to some extent to the occurrence of abnormal VM (21). However, it often persists after antibiotic treatment. This could mean that it easily tolerates the presence of other bacteria, or that it helps to restore a Lactobacilli-dominated VM during and after dysbiosis and/or antibiotic treatment. L. iners has been reported to become a predominant part of the microbial community when the VM transitions between abnormal and normal states (22). Consistent with previous study (23), anaerobic L. iners had a significantly lower diversity in persistent group than the other two. It is inferred that the role of the L. iners in reconstruction the Lactobacilli-dominated VM was eliminated for the low abundance, thus leading to the persistent of HR-HPV infection.
P. bivia (previously called Bacteroides) and P. disiens are two very common members of the nonpigmented Prevotella, both important in obstetric and gynecologic infections. P. disiens is commonly isolated from the urogenital tract (24), and occasionally isolated from polymicrobial infections of the upper respiratory tract (25), central nervous system (26), urogenital (27) and oral tract (28). The relative abundance of P. disiens in group A was lower than that in group C, but higher than that in group B. However, the relative abundance of P. bivia was significantly higher in group A than B and C. Thus, we infer that P. bivia and P. disiens may play certain roles in HPV persistence. Few articles could be found on the relationship between Enterococcus durans, Porphyromonas uenonis and HPV infection. In this study, we found that the relative abundance of obligately anaerobic Porphyromonas uenonis and Enterococcus durans were significantly higher in group A than group B and C, which is also the most important finding of our study.
In this pilot study, we explored a range of bacteria that may be related to HPV persistent infection. According to the guidelines (29) if a specimen was positive for HPV-16/18, coloscopy was performed. However, for those whose specimen was positive for the other 12 types HR-HPV, the Pap testing is a useful stratification algorithm. With this being the case, if the result of Pap testing is normal, the patients will go for coloscopy only if the other 12 types HR-HPV persist for at least 1 year. It is worth noticing that the limitations of Pap testing include specimen adequacy for evaluation and variable sensitivity (30). Thus, we analyzed the potential bacteria biomarker to see if they can act as an auxiliary stratifying measure. For the patients whose cervical Pap smear was normal, our results showed that there are 11.11% (3/27) confirmed as CIN2+ in persistent other 12 types of HR-HPV infection group, while 2.22% in incident HPV infection group. In this situation, many of the patients underwent invasive examination and overtreatment. If the vaginal microorganism biomarkers were combined, more cases may avoid this. With the significantly different VM, we performed the ROC curve to find the cut-off relative abundance of potential microbiome biomarker to help predicting the appearance of CIN 2+, aiming at decrease the occurrence of invasive inspection. The specificity can reach to 66.67% and the sensitivity is still 100% when the relative abundance of P. bivia is over 0.05554% with Prevotella disiens being under 0.02196%. The potential VM biomarker could be used to facilitate the clinical practice. This finding in our study is of great importance.
At present, HPV vaccines are the main prevention strategy for cervical cancer. Our findings suggest that specific VM may be involved in HR-HPV infection persistence, and even involved in the pathogenesis of CIN. Probiotics have been used in a similar manner to reduce the recurrence of bacterial vaginitis (BV), through accurate, targeted modification of the bacterial community (31). This study may provide new assistant indicators for predicting the appearance of CIN2+ and new ideas for the treatment of vaginal dysbiosis related persistent HPV infection in the future. Modulation of the VM with oral or vaginal regimes to a Lactobacillus spp.-dominant microbiome may be able to promote HPV clearance or even reverse the process of tumor-genesis (31,32). Microbiome modulation could also represent low-cost future therapeutic strategies.
The strength of this study is that we found the significantly different biomarkers assisting in predicting of the HR-HPV persistent infection. What’s more important, the relative abundance of P. bivia being over 0.05554% with Prevotella disiens being under 0.02196% can assist predicting the appearance of CIN2+. However, there are also several limitations in this study. Given that this was a cross-sectional study, we were unable to determine the causal relation between VM and HPV infection. Further research is required to understand the factors promoting persistence as well those triggering carcinogenetic pathways. Further longitudinal study is needed to investigate the changes and stability of VM during transition from acute HPV infection to persistent infection, and through to development of CIN and cancer. An ongoing validation should be performed in a large sample of the role of P. bivia and Prevotella disiens in prediction of appearance of CIN2+.
Conclusions
In conclusion, vaginal dysbiosis likely is a largely understudied yet important risk factor in HPV and cervical cancer epidemiology. Microbiome modulation may promote the clearance of HPV and reverse the natural history of HPV infection. An understanding of the functional properties of the VM is required in order to complement what we already know about their structure.
Supplementary
DNA extraction and the amplification of bacterial 16S rRNA V4 gene region and Illumina sequence were showed in the supplementary material
DNA extraction
Total genomic DNA was extracted from cervical samples using CTAB method. One thousand µL CTAB lysate was pipetted into 2.0 mL EP tube, and 20 µL lysozyme and then appropriate amount of sample was added to the lysate. The mixture was incubated for 1.5 h at 65 °C, during which the sample should be inverted several times to mix fully so that it can be fully lysed. After centrifugation, 950 µL supernatant was mixed with equal volume phenol (pH 8): chloroform: isoamyl alcohol (25:24:1), and then centrifugated at 12,000 rpm for 10 min. Supernatant was again mixed with equal volume chloroform: isoamyl alcohol (24:1), and centrifugated at 12,000 rpm for 10 min. Supernatant was pipetted into 1.5 mL centrifuge tube, and mixed with 3/4 volume of isopropanol, and precipitated at −20 °C. After centrifuging, the liquid was washed with 1 mL 75% ethanol twice, and then dried in ultra clean workbench or room temperature. The DNA sample was dissolved in 51 µL ddH2O, and hatched at 55–60 °C for 10 min for solubilizing if necessary. One µL RNase A was added to digest RNA, and the DNA sample was placed at 37 °C for 15 min. DNA concentration and purity was monitored on agarose gels at 1%. According to the concentration, DNA was diluted to 1 ng/µL using sterile water.
Amplification of bacterial 16S rRNA V4 gene region and Illumina sequence
Using extracted genome DNA as template, the V4 region of the bacterial 16S rRNA gene was PCR-amplified using the primers 515F (5'-GTGCCAGCMGCCGCGGTAA-3') and 806R (5'-GGACTACHVGGGTWTCTAAT-3') with the barcode. All PCR reactions were carried out with Phusion® High-Fidelity PCR Master Mix with GC Buffer (New England Biolabs), using 25 µL Taq PCR mix (2×), 1 µL 10 µM primer F, 1 µL 10 µM primer FR, 2.5 µL gDNA, 8.0 µL H2O. An initial denaturation step of 95 °C for 5 min was carried out, followed by 34 cycles of denaturation (94 °C, 1 min), annealing (57 °C, 45 s) and extension (72 °C, 1 min), and a final elongation step of 10 min at 72 °C,cooling for 5 min at 16 °C. Mix same volume of 1× loading buffer (contained SYBR green) with PCR products and operate electrophoresis on 2% agarose gel for detection. Samples with bright main strip between 400–450 bp were chosen for further experiments. PCR products was mixed in equidensity ratios. Then, mixture PCR products was purified with Qiagen Gel Extraction Kit (Qiagen, Germany). Sequencing libraries were generated using TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, USA) following manufacturer’s recommendations and index codes were added. The library quality was assessed on the Qubit@ 2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. At last, the library was sequenced on an Illumina HiSeq. 2500 platform and 250 bp paired-end reads were generated.
Data analysis
The original data were pretreated by BIPES data analysis process. In total 26,791,191 reads were obtained from 329 samples with an average number of reads per sample of 81,432 reads. Sequences analysis were performed by Uparse software (Uparse v7.0.1001). Sequences with a 97% similarity threshold were assigned to the same operational taxonomic units (OTUs). Representative sequence for each OTU was screened for further annotation. Species annotation was analyzed using the Mothur method and the SSUrRNA database of SILVA (threshold value 0.8–1). Multiple sequence alignment was conducted using the MUSCLE software (Version 3.8.31) to study the phylogenetic relationship of different OTUs and the difference of the dominant species in two groups. The subsequent analysis includes Alpha and Beta diversity analysis. Alpha diversity is applied in analyzing complexity of species diversity for a sample through 6 indices, including Observed-species, Chao 1, Shannon, Simpson, ACE, Good-coverage. All these indices in our samples were calculated with QIIME software (Version 1.7.0) and displayed with R software (Version 2.15.3). Chao 1 and ACE were selected to identify Community richness. Shannon and Simpson were used to identify Community diversity. And coverage was used to characterize Sequencing depth. Beta diversity analysis was used to evaluate differences of samples in species complexity. Based on the unweighted_unifrac distance, using QIIME software (Version 1.7.0) to carry out the analysis of principal component of vaginal microbiota. Cluster analysis was preceded by principal component analysis (PCA), which was applied to reduce the dimension of the original variables using the FactoMineR package and ggplot2 package in R software (Version 2.15.3). Principal coordinate analysis (PCoA) was performed to get principal coordinates and visualize from complex, multidimensional data. A distance matrix of weighted or unweighted UniFrac among samples obtained before was transformed to a new set of orthogonal axes, by which the maximum variation factor is demonstrated by first principal coordinate, and the second maximum one by the second principal coordinate, and so on. PCoA analysis was displayed by WGCNA package, stat packages and ggplot2 package in R software (Version 2.15.3). Unweighted Pair-group Method with Arithmetic Means (UPGMA) Clustering was performed as a type of hierarchical clustering method to interpret the distance matrix using average linkage and was conducted by QIIME software (Version 1.7.0). Using linear discriminant analysis (LDA) coupled with effect size measurements (LEfSe) to analyze the difference of structure and composition of vaginal microbial communities between two groups.
Acknowledgments
We thank all the participants of the study.
Funding: This study was funded by CAMS Youth Talent Award Project (No. 2018RC320006), CAMS Innovation Fund for Medical Sciences (CIFMS) (No. 2016-I2M-1-002), and Chinese Academy of Medical Sciences Initiative for Innovative Medicine (CAMS-2017-I2M-1-002).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Ethical approval was obtained from the Ethics Committee of Peking Union Medical College Hospital (PUMCH), Beijing, China (No. JS-1634). All experiments were performed in accordance with relevant guidelines and regulations. The registration No. in clinicaltrials.gov is NCT03548740. Written informed consent was obtained from all participants.
References
- Chesson HW, Dunne EF, Hariri S, et al. The estimated lifetime probability of acquiring human papillomavirus in the United States. Sex Transm Dis 2014;41:660-4. [Crossref] [PubMed]
- Rodríguez AC, Schiffman M, Herrero R, et al. Rapid clearance of human papillomavirus and implications for clinical focus on persistent infections. J Natl Cancer Inst 2008;100:513-7. [Crossref] [PubMed]
- Shanmugasundaram S, You J. Targeting Persistent Human Papillomavirus Infection. Viruses 2017. [Crossref] [PubMed]
- Elfgren K, Jacobs M, Walboomers JM, et al. Rate of human papillomavirus clearance after treatment of cervical intraepithelial neoplasia. Obstet Gynecol 2002;100:965-71. [PubMed]
- Kyrgiou M, Mitra A, Moscicki AB. Does the vaginal microbiota play a role in the development of cervical cancer? Transl Res 2017;179:168-82. [Crossref] [PubMed]
- Mitra A, Macintyre DA, Marchesi JR, et al. The vaginal microbiota, human papillomavirus infection and cervical intraepithelial neoplasia: what do we know and where are we going next? Microbiome 2016;4:58. [Crossref] [PubMed]
- Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 2011;108 Suppl 1:4680-7. [Crossref] [PubMed]
- Liu MB, Xu SR, He Y, et al. Diverse Vaginal Microbiomes in Reproductive-Age Women with Vulvovaginal Candidiasis. Plos One 2013;8:e79812. [Crossref] [PubMed]
- Borgdorff H, Armstrong SD, Tytgat HL, et al. Unique Insights in the Cervicovaginal Lactobacillus iners and L. crispatus Proteomes and Their Associations with Microbiota Dysbiosis. PLoS One 2016;11:e0150767. [Crossref] [PubMed]
- Smith SB, Ravel J. The vaginal microbiota, host defence and reproductive physiology. J Physiol 2017;595:451-63. [Crossref] [PubMed]
- Brusselaers N, Shrestha S, van de Wijgert J, et al. Vaginal dysbiosis and the risk of human papillomavirus and cervical cancer: systematic review and meta-analysis. Am J Obstet Gynecol 2019;221:9-18.e8. [Crossref] [PubMed]
- Shoskes DA, Shahed AR. Detection of bacterial signal by 16S rRNA polymerase chain reaction in expressed prostatic secretions predicts response to antibiotic therapy in men with chronic pelvic pain syndrome. Tech Urol 2000;6:240-2. [PubMed]
- Castle PE, Rodriguez AC, Burk RD, et al. Long-term persistence of prevalently detected human papillomavirus infections in the absence of detectable cervical precancer and cancer. J Infect Dis 2011;203:814-22. [Crossref] [PubMed]
- Gajer P, Brotman RM, Bai G, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med 2012;4:132ra152. [Crossref] [PubMed]
- Jouseimies-Somer H, Summanen P, Citron D, et al. Wadsworth-KTL anaerobic bacteriology manual. 6th edition. Belmont: Star Publishing Company, 2002.
- Olsen I, Shah HN, Gharbia SE. Taxonomy and biochemical characteristics of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. Periodontol 2000 1999;20:14-52. [Crossref] [PubMed]
- Li J, McCormick J, Bocking A, et al. Importance of vaginal microbes in reproductive health. Reprod Sci 2012;19:235-42. [Crossref] [PubMed]
- Macklaim JM, Fernandes AD, Di Bella JM, et al. Comparative meta-RNA-seq of the vaginal microbiota and differential expression by Lactobacillus iners in health and dysbiosis. Microbiome 2013;1:12. [Crossref] [PubMed]
- McMillan A, Dell M, Zellar MP, et al. Disruption of urogenital biofilms by lactobacilli. Colloids Surf B Biointerfaces 2011;86:58-64. [Crossref] [PubMed]
- Aroutcheva A, Gariti D, Simon M, et al. Defense factors of vaginal lactobacilli. Am J Obstet Gynecol 2001;185:375-9. [Crossref] [PubMed]
- Verstraelen H, Verhelst R, Claeys G, et al. Longitudinal analysis of the vaginal microflora in pregnancy suggests that L. crispatus promotes the stability of the normal vaginal microflora and that L. gasseri and/or L. iners are more conducive to the occurrence of abnormal vaginal microflora. BMC Microbiol 2009;9:116. [Crossref] [PubMed]
- Jakobsson T, Forsum U. Lactobacillus iners: a marker of changes in the vaginal flora? J Clin Microbiol 2007;45:3145. [Crossref] [PubMed]
- Lee JE, Lee S, Lee H, et al. Association of the vaginal microbiota with human papillomavirus infection in a Korean twin cohort. PLoS One 2013;8:e63514. [Crossref] [PubMed]
- Fountoukis T, Tsatsanidis N, Tilkeridou M, et al. Abdominal rectus muscle pyomyositis: Report of a case and review of the literature. Infect Dis Rep 2018;10:7522. [Crossref] [PubMed]
- Gebhardt B, Herrmann K, Roessner A, et al. Laryngorhinootologie 2010;89:266-9. [Differential diagnosis of unilateral necrotic tonsillitis]. [Crossref] [PubMed]
- Takahashi K, Hasegawa Y, Nishimoto Y, et al. Solitary actinomycotic brain abscess: case report. Brain Nerve 2012;64:689-95. [PubMed]
- Persson R, Hitti J, Verhelst R, et al. The vaginal microflora in relation to gingivitis. BMC Infect Dis 2009;9:6. [Crossref] [PubMed]
- Salari MH, Kadkhoda Z. Rate of cultivable subgingival periodontopathogenic bacteria in chronic periodontitis. J Oral Sci 2004;46:157-61. [Crossref] [PubMed]
- Huh WK, Ault KA, Chelmow D, et al. Use of primary high-risk human papillomavirus testing for cervical cancer screening: interim clinical guidance. Obstet Gynecol 2015;125:330-7. [Crossref] [PubMed]
- Hutter JN, Decker CF. Human papillomavirus infection. Dis Mon 2016;62:294-300. [Crossref] [PubMed]
- Vujic G, Jajac Knez A, Despot Stefanovic V, et al. Efficacy of orally applied probiotic capsules for bacterial vaginosis and other vaginal infections: a double-blind, randomized, placebo-controlled study. Eur J Obstet Gynecol Reprod Biol 2013;168:75-9. [Crossref] [PubMed]
- Kyrgiou M, Koliopoulos G, Martin-Hirsch P, et al. Obstetric outcomes after conservative treatment for intraepithelial or early invasive cervical lesions: systematic review and meta-analysis. Lancet 2006;367:489-98. [Crossref] [PubMed]


