
Exploration of the immune-related signature and immune infiltration analysis for breast ductal and lobular carcinoma
Introduction
Breast cancer (BC) has gradually become a commonly diagnosed cancer type worldwide. According to Globocan (1), there are 2,088,849 women who are diagnosed with BC and 626,679 patients who die from it per year; BC has the highest incidence/mortality among female malignant tumour types. Surgical and drug therapies are considered the most common treatments for BC. However, both methods result in physical injury, coupled with poor prognosis and high recurrence (2,3). With the accumulation of molecular research on BC, targeted therapy has gradually become a key means of treatment (4). Trastuzumab (5), temsirolimus (6) and bevacizumab (7) are common targeted drugs in the clinic. There are abundant clinical studies on differential gene expression in BC cases, which could provide accurate differential gene data. This could supply the theoretical basis for BC targeted therapy research and unearth novel targets for clinical diagnosis, treatment and prognosis of BC.
The immune response is considered an important pathway in the occurrence and prognosis of BC, which means that the enhancement of the tumour immune response could provide an effective benefit for those with BC, improving clinical results and overall survival (OS) (8). Immunotherapies such as the anti-growth factor 2 (HER2) Th1 response are becoming a favourable treatment approach for BC. For example, by blocking HER2, the Th1 response was inhibited, and 40–67% of patients achieved a pathologic complete response (pCR) (9). Another study discovered the importance of immune escape in tumour cell chemotherapy resistance, where programmed death-1 (PD-1)/programmed death ligand 1 (PD-L1) inhibitors could be regarded as a potential approach in the triple-negative breast cancer (TNBC) (10). Studies have shown that the components of the tumour immune environment, especially the tumour-infiltrating lymphocytes (TIL) communities, are correlated with patients’ disease-free survival (DFS) and OS (11). However, the previous investigations mainly focused on the different gene expression (DGE) or the tumour immune environment of BC. The biomarkers of tumour immune cells for BC are still poorly understood, and feasible immune biomarkers for the prediction of BC patients’ prognosis are lacking. Moreover, as Egelston’s research has shown (12), although TIL can be regarded as the immune signature of the tumour, the complex function of different cell subsets in TIL limits its utilization in BC research. Therefore, it is essential to confirm the relationship of specific biomarkers of tumour immune cells and BC and find a robust immune target that can serve as a perspective of tumour immunology and may become a target for immunotherapy of BC.
In this study, we utilized the transcriptome data from The Cancer Genome Atlas (TCGA) to develop an immune-related risk signature for BC, and we validated 1,399 immune-related genes which correlated to the OS of BC. To evaluate the clinical value of these signatures, the TAIG risk model was calculated based on the multivariate Cox regression results, and we conducted the correlation analysis between TAIG and the clinical characteristics. To further investigate the molecular and immune profiling of the signature, we evaluated the associations between the immune signature and several immune cells based on TIMER.
Methods
Data acquisition and processing
We downloaded the genome expression data of 1,178 samples from the TCGA database (https://portal.gdc.cancer.gov/), including 1,066 tumour samples and 112 matched normal samples. Besides, we collected a total of 1,094 patients from the METABRIC cohort with complete survival information and expression data. Meanwhile, we obtained a list of immune related signatures from the InnateDB database (https://www.innatedb.ca/), which provides available resources for immunology research. Then, limma package was utilized to determine the normalization of gene expression, and we conducted a differential analysis to screen the abnormally expressed genes in tumour versus normal samples. We then obtained the intersect immune signature on a Venn diagram. To investigate the biological function of differential immune genes in BC, we transferred the gene symbol with Entrez ID and performed the Gene Ontology (GO) and KEGG analyses via the clusterProfiler, org.Hs.eg.db, enrichplot and ggplot2 packages. The significantly enriched GO items and signalling pathways are shown using bar plots and dot plots. Meanwhile, we also obtained the clinical information of patients from the database via the TCGAbiolinks package, including age, gender, TNM stages, tumour grades, and follow-up with vital status.
Construction of the TAIG risk score in BC
Since we screened the differentially expressed immune genes, we further investigated the hub immune genes for predicting the prognosis of BC. First, we selected the differential immune genes and conducted the univariate Cox regression analysis to find the prognostic signature among them. Then, we chose the significant signatures with P<0.001 and prepared a Least Absolute Shrinkage And Selection Operator (LASSO) model to reduce the variables. Afterwards, we conducted a multivariate Cox analysis to construct the TAIG (tumor-associated immune genes) risk model calculated as: TAIG = Ʃ (βi × Expi), where βi, the coefficients, represented the weight of the respective signature and Expi represented the expression value. Accordingly, we could calculate the TAIG risk score for each patient and classified patients into a high- or low-TAIG group with the median value as the cut-off data. The distribution of the vital status was illustrated according to the TAIG risk score levels. We also showed the differentially expressed levels of immune signatures in two TAIG groups via a pheatmap package.
Assessment of TAIG and associations with clinical variables
Given the identification of hub immune signatures associated with survival outcomes in BC, we intended to explore the predictive value of TAIG. We utilized the timeROC package to conduct the receiver operating characteristic (ROC) curve to show the 5-year OS prediction. Meanwhile, a Kaplan-Meier analysis with a log-rank test was used to assess the survival difference between low- and high-TAIG groups. Moreover, we discovered the underlying relationships between TAIG and several clinical features consisting of TNM stages or pathological stages. The Wilcoxon rank-sum test was utilized for comparing differential levels of TAIG between two groups, but the Kruskal-Wallis test was appropriate when dealing with three or more groups.
Explorations of associations between TAIG and immune infiltrates
TIMER is a comprehensive resource for systematic analysis of immune infiltrates across multiple malignancies. The abundances of six immune infiltrates (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages and dendritic cells) are estimated by the statistical method, which is validated by pathological estimations. This web server allows investigators to input function-specific parameters, with resulting figures dynamically displayed to conveniently access the tumour’s immunological, clinical, and genomic features. Based on the public resources, we could discover the identified immune signature with tumour-infiltrating immune cells. Pearson’s correlation coefficient and the estimated P value were calculated to evaluate the associations between TAIG and immune infiltration cells.
CIBERSORT and differential abundance of immune cells in two TAIG groups
Given the important roles of tumour-infiltrating immune cells in the tumour microenvironment, we utilized CIBERSORT, a newly developed deconvolution algorithm, to determine the fractions of 22 immune cells in each sample. We exhibited the results as a box plot, and the various immune cells were annotated under the legend. Furthermore, we conducted a Wilcoxon rank-sum test to compare the differential abundance of immune cells in the two TAIG groups. Last, we illustrated the differential density of immune cells in two TAIG groups using a pheatmap package, where the colours ranging from green to red represented the low to high infiltrating levels.
Statistical analysis
The Student’s t-test was suitable for continuous variables, while categorical variables were compared by a Chi-square (χ2) test. A Wilcoxon rank-sum test was a non-parametric statistical test mainly utilized for comparing two groups, and a Kruskal-Wallis test was used for two or more groups. Differential analysis and normalization were mainly conducted using the “limma” package. The Cox regression model or Kaplan-Meier analysis with a log-rank test was performed using the “survival” package. All statistical analyses were performed using R software (Version 3.5.2), and a P value <0.05 was considered to be statistically significant.
Results
Identification of prognostic immune-related signatures in BC
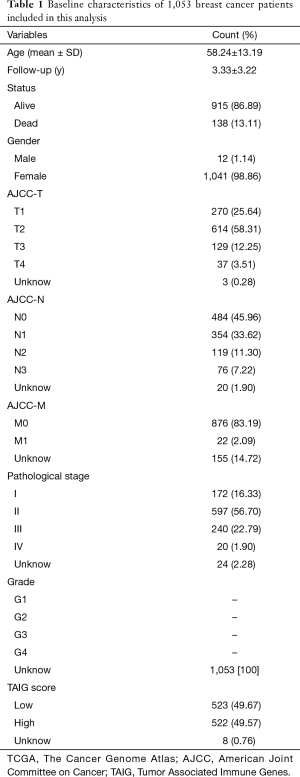
We obtained 1,178 samples from the TCGA database, consisting of 1,066 tumour samples and 112 corresponding normal samples. We excluded the patients with insufficient clinical data, and the average age was 58.24±13.19 among the 1,053 tumour samples. The complete information is shown in Table 1. Meanwhile, the corresponding clinical information of METABRIC patients was collected in http://cdn.amegroups.cn/static/application/64f39528984716b90837edd92e9d54e9/10.21037atm.2019.11.11-1.xlsx. We found 1,045 tumour samples that were matched with complete survival and transcriptome data and were included in our subsequent study. We used the limma package to conduct the normalization of transcriptome profiles and screened approximately 10,563 differentially expressed genes with |log fold change| >1 and false discovery rate (FDR) <0.05. The volcano plot is shown in Figure 1 to exhibit the up-regulated or down-regulated genes. We downloaded a list of 4,678 immune-related genes from the InnateDB database to intersect approximately 1,399 differential immune-related signatures, which is shown in a Venn diagram (Figure 1). The functional enrichment analysis revealed that these differentially expressed immune genes participated in several areas of immune-related crosstalk, including cytokine-cytokine receptor interactions, Th1 and Th2 cell differentiation and the JAK-STAT signalling pathway.
Full table
Establishment of TAIG risk score and model assessment
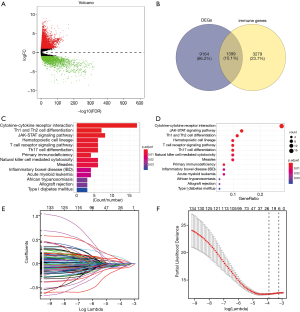
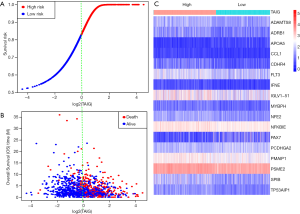
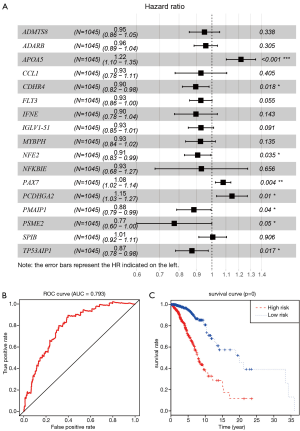
Given the fact that these immune related signatures showed differential levels in tumour versus normal samples, we further wanted to screen the prognostic genes among them. First, we merged the expression profiles of differential immune genes with survival data and utilized the univariate Cox analysis to obtain a list of 136 prognostic signature with P<0.01. Then, we selected 136 genes with which to perform the LASSO regression method to find the most significantly prognostic immune genes. As the screening procedure shows in Figure 1, we identified the 17 hub immune genes shown in Table 2. Then, we performed a multivariate Cox regression analysis to calculate each coefficient of signature, which represented the weight of each gene. The TAIG risk score was accordingly calculated as: TAIG =−0.050499× ADAMTS8 −0.041650× ADRB1 +0.197286× APOA5 −0.074406× CCL1 −0.109735× CDHR4 −0.074999× FLT3 −0.108292× IFNE −0.074270× IGLV1-51 −0.077379× MYBPH −0.096452× NFE2 −0.071412× NFKBIE +0.076329×PAX7 +0.136178× PCDHGA2 −0.122293× SPIB −0.135507× TP53AIP1. We thus could classify the 1045 BC patients into 523 low-TAIG and 522 high-TAIG groups. We observed that the patients with a high TAIG risk score suffered more survival risks, as shown in Figure 2. Moreover, the differentially expressed levels of hub genes in the two TAIG groups are shown by the heatmap in Figure 2. What is more, the AUC of the ROC plot in Figure 3 was 0.793, indicating the superior predictive accuracy of the TAIG risk score for the prognosis of BC. The Kaplan-Meier analysis with the log-rank test also further demonstrated that patients with high TAIG scores were associated with poor OS (P<0.001) in Figure 3. In addition, the correlation analysis between TAIG levels and clinical characteristics also revealed that high TAIG levels correlated with high AJCC-TNM stages and advanced pathological stages (P<0.01), implicating the significant clinical value of TAIG. Last, we further validated the TAIG in an independent data set in METABRIC cohort and found that the 3-, 5- and 7-year AUC in predicting OS was 0.732, 0.784 and 0.829, respectively (Figure 4). Correspondingly, the Kaplan-Meier curve in 1,094 patients showed that the higher TAIG risk scores indicated poor survival outcomes with P<0.0001 (Figure 4).

Full table
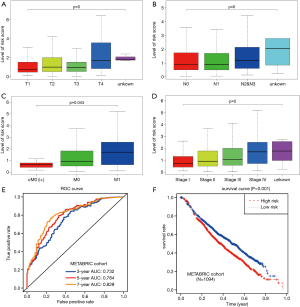
Associations of TAIG with tumour-infiltrating immune cells
Given the important roles of infiltrating immune cells in the tumour microenvironment, we integrated the comprehensive analysis of immune signatures combined with immune infiltrates. Based on the TIMER database, we discovered the relationships between 17 hub immune signatures and tumour purity or several important immune cells. We observed that most of these signatures were associated with immune cells, especially PCDHGA2, SPIB, ADRB1, FLT3, and NFKBIE, which are shown in Figure 5.
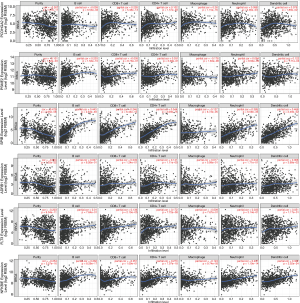
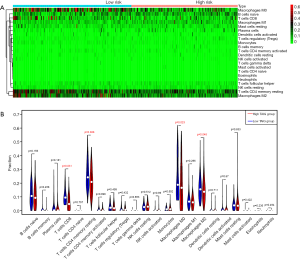
Moreover, we further utilized the CIBERSORT algorithm to determine the estimated fractions of 22 immune cells in each sample. We excluded the samples with a calculated P value >0.05 to guarantee the accuracy of the analysis, and we illustrated the results in a box plot, where the differential immune cells were annotated with various colours and the sum of immune fractions in each sample was equal to one (Figure 6).
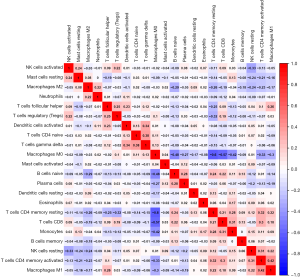
Differential abundance of tumour-infiltrating immune cells in two TAIG groups
Since we analysed the potential associations of immune signature with several immune cells, we speculated that there were differential distributions of immune cells in the two TAIG groups (Figure S1). The heatmap exhibited the infiltrating difference of immune cells in two groups where the colours ranging from green to red represented the infiltration density from low to high (Figure 7). Additionally, we utilized the Wilcoxon rank-sum test to accurately compare the difference and found that several immune cells conferred significantly lower infiltrating density in the high TAIG groups, including CD8+ T cells (P=0.031), memory resting CD4+ T cells (P=0.026), M0 macrophages (P=0.023), as well as M2 macrophages (P=0.048). Several previous studies have already demonstrated that lower immune infiltration in the tumour microenvironment likely results in a poor prognosis in BC. Based on the integrative analysis, we inferred that the risk factor of TAIG was associated with lower immune infiltrates, thus leading to worse survival outcomes.
Discussion
Although both the DEGs and the immune environment of BC have been well studied, considering that BC has the highest incidence/mortality among female malignant tumours types, the immune biomarkers for the prediction of the tumour, especially in the TIL communities, are lacking. Taking into account the importance of immune infiltration in the development and proliferation of cancer, the finding of novel BC immune-related biomarkers may prove useful for immunotherapy. The present study established a robust immune-related risk signature for BC using the TCGA datasets. With the adjustment of the TAIG risk model, these signatures were able to predict the patients’ OS. In addition, we found a significant correlation of these signatures to immune cells according to the TIMER datasets. BC patients with a high immune risk of the selected signatures can have a large deviation in the inflammatory microenvironment. Finally, 17 immune-related signatures were selected as the hub immune genes. According to the KEGG and GO functional results, our signatures take part in cytokine-cytokine receptor interactions, Th1 and Th2 cell differentiation and the JAK-STAT signalling pathway. This can further explain the effect of these signatures on the relative cytokines and their receptors in the pathogenesis of cancer (5). Taken together, a higher TAIG levels means a poorer OS. The BC patients with high immune risk levels could stimulate the deterioration of the tumour and accelerate the progression of BC, leading to poor OS.
To determine the clinical values of these signature, the associations among TAIG risk level, AJCC-TNM stages, and patients’ OS were evaluated. Patients with a high immune risk score tended to have high AJCC-TNM stages and advanced pathological stages. Our investigation was the first to uncover the immune-related differences between the clinical stages of BC. Based on the results, the high immune risk microenvironment may promote the development and recurrence of BC, which leads to an advanced stage and relapse of the tumour. Therefore, our signatures can also be regarded as predictors of the development, recurrence and survival outcomes of BC. The multivariate analysis result indicated that the immune-related risk signatures cannot only be independent predictors of BC in the clinic but also potential immunotherapy targets for BC patients in the future.
According to the results, 17 hub immune signatures with tumour purity, or several important immune cells, and PCDHGA2, SPIB, ADRB1, FLT3, and NFKBIE were the most significant factors. PCDH is expressed in the nervous system mainly, and PCDHγ participates in synaptogenesis and mediates the connection between astrocytes and neurons (13). PCDHGA2 belongs to the PCDHγ family and regulates the function of the nervous system, such as synaptic generation (14) and neuronal growth, maturation and differentiation (15), among cells in the intracellular region. PCDHGA2 mutations can be regarded as the signature correlated with several cancer types. In Ping’s research (16), high-recurrent-risk patients of invasive lobular cancer of the breast tended to have PCDHGA2 mutations. Song’s research demonstrated that (17), in aggressive papillary thyroid microcarcinomas patients, PCDHGA2 can promote the aggressiveness of the tumour. The overexpression of PCDHGA2 mutations could accelerate the metastasis of the tumour by its adhesion-induced function. Therefore, we suggest that PCDHGA2 is a bridge between neuroscience and immune-oncology.
Because of the conserved ETS domains, the ETS family could induce specific DNA binding. SPIB is a transcription factor of the ETS family that is homologous with the SPI1 (PU.1) gene in sequence. In a previous study, SPIB participated in the growth of B lymphocyte and plasmacytoid dendritic cells (pDCs), thus regulating the immune function of these cells (18). Considering the immunomodulatory role of SPIB in B lymphocytes, the copy amplification, translocation and chromosome mutations of SPIB can promote tumour-immune microenvironment remodelling (19,20). There have been studies showing that SPIB regulates the infiltration of blood cells, which was one of the suggested mechanisms for several leukaemias, such as acute lymphoblastic leukaemia (21) and acute myeloid leukaemia (22) Moreover, SPIB is also overexpressed in hepatoma and lung carcinoma tissue (23). Regarding its immunologic function in B-lymphocytes, we suggest that SPIB is widely involved in the invasion and metastasis of cancer.
Similar to SPIB, FLT3 is another important signature in haematological diseases. In myeloid cells, FLT3 is expressed restrictedly in CD34+ cells (24). FLT3 and its ligand (FLT3-L), assembled by marrow stroma cells, play an important role in self-renewal, proliferation and differentiation of hematopoietic stem cells. Therefore, FLT3 is a critical factor for leukaemia patients. FLT3 internal tandem duplication (FLT3-ITD) mutations are associated with poor survival outcomes of AML (25). Furthermore, in the ALL patients, FLT3 mutations usually led to an increase in lymphoid markers such as CD7, CD13, CD34, CD117 (26). Hence, we consider that FLT3 connected the lymphoblast immunity reaction and the immunological microenvironment. In our study, FLT3 is overexpressed in CD7 and CD13 of BC patients with high immune risk. These findings suggest that tumour microenvironment remodelling caused by immunomodulatory dysfunction can promote the expression of FLT3 in BC patients.
Most prior studies support a dominant role of the NFKB pathway in BC, which was one of the multiple mechanisms of immune regulation to restrain immune responses and regulate their propagation (27). The inhibition of p53 transcription resulting from NFKB pathway activation could limit the transcription of other apoptosis-promoting genes, thus inducing the proliferation of tumour cells. Hence, the blockade of the NFKB pathway can be regarded as an immune approach in BC immunotherapy (28). As an important inhibitor of the NFKB pathway, NFKBIE were strongly associated with chronic lymphocytic leukaemia (CLL) and melanoma, where NFKBIE mutations occur in 7% of CLL patients with a poor prognosis (29) and 14.5% of melanoma patients with a poor prognosis (30). Additionally, NFKBIE regions in granulocytes and macrophages participate in the regulation of the autoimmune response, which was an important immune signature in the rheumatoid arthritis microenvironment (31). These studies show that NFKBIE can limit tumour trans-differentiation from immune infiltration by the NFKB pathway.
Furthermore, we tried to investigate the correlation of these immune signatures and patient prognosis. Further studies showed that immune cells had a significantly lower infiltrating density in high TAIG groups, including CD8+ T cells (P=0.031), memory resting CD4+ T cells (P=0.026), M0 macrophages (P=0.023), and M2 macrophages (P=0.048). This is in agreement with the study of Tawfik (32) and Bense (33): the decrease of immune infiltrating cells in the tumour microenvironment may be related to poor prognosis in BC. In our study, high-TAIG group patients had less CD8+ and CD4+ T-cell infiltration, leading to the poor prognosis of the patients. Nevertheless, further investigations are needed to confirm the mechanism of these signatures in the immune microenvironment.Taken together, our study is the first to identify and validate 17 immune-related genes according to the TAIG scoring system, and our results can further prove a connection between the TAIG score and the prognosis of BC patients, which could indicate the immune infiltration intensity in the BC microenvironment. These selected signatures can also provide novel immune targets for BC immune-related treatment in the future.
Compared with the traditional study of BC bio-markers, our study analysed many clinical samples in the public database and focused on the expression of immune-related genes in immune cells. With the study of the characteristics of cellular immune infiltration, the relationship between the regulation of immune-related genes and the incidence and prognosis of BC was investigated in depth. The use of CIBERSORT also greatly improved the efficiency of research and avoided the heavy experimental process. This study used a large number of queues in the TCGA database to identify and validate immune markers and to further assess their clinical risks. However, because this study entirely relies on fitting, its clinical accuracy needs to be further assessed in cohort studies. In addition, the markers obtained in this study need to be combined with clinical studies to determine their functions and provide a basis for future clinical research.
With the method proposed in this study, we can utilize public databases for immune inflammation studies without the need for physical pathways or understanding the potential cell composition of samples. The method proposed in this study enables us to fully examine the immune inflammation patterns of BC mentioned in previous studies, to assess the specific distribution of immune cells and the changes of corresponding biomarkers, and to discover new biological relationships among them. The method proposed in this study provides a new therapeutic target for clinical treatment. The feasibility of this study is high, provides a feasible method for future BC immune research and can be extended to other cancer research.
Acknowledgments
Funding: The Project was supported by the Open fund of Key Laboratory of Ministry of Education for TCM Viscera-State Theory and Applications, Liaoning University of Traditional Chinese Medicine. The Project was supported by the Shenyang Science and Technology Project (18-013-0-44). The Project was supported by the Natural Science Fund Project of Science and Technology Department of Liaoning Province (No. 0180550645). The Project was supported by Research Administration Office of Liaoning Economy Vocational and Technical College (Ljz2018-qn-07).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Maishman T, Cutress R, Hernandez A, et al. Local Recurrence and Breast Oncological Surgery in Young Women With Breast Cancer: The POSH Observational Cohort Study. Ann Surg 2017;266:165-72. [Crossref] [PubMed]
- van Ramshorst MS, van Werkhoven E, Mandjes IAM, et al. Trastuzumab in combination with weekly paclitaxel and carboplatin as neo-adjuvant treatment for HER2-positive breast cancer: The TRAIN-study. Eur J Cancer 2017;74:47-54. [Crossref] [PubMed]
- The Lancet. Breast cancer targeted therapy: successes and challenges. Lancet 2017;389:2350. [Crossref]
- Inoue K, Ninomiya J, Saito T, et al. Correction to: Eribulin, trastuzumab, and pertuzumab as first-line therapy for patients with HER2-positive metastatic breast cancer: a phase II, multicenter, collaborative, open-label, single-arm clinical trial. Invest New Drugs 2019;37:592-3. [Crossref] [PubMed]
- Boers-Sonderen MJ, de Geus-Oei LF, Desar IM, et al. Temsirolimus and pegylated liposomal doxorubicin (PLD) combination therapy in breast, endometrial, and ovarian cancer: phase Ib results and prediction of clinical outcome with FDG-PET/CT. Target Oncol 2014;9:339-47. [Crossref] [PubMed]
- Nahleh Z, Botrus G, Dwivedi A, et al. Bevacizumab in the neoadjuvant treatment of human epidermal growth factor receptor 2-negative breast cancer: A meta-analysis of randomized controlled trials. Mol Clin Oncol 2019;10:357-65. [PubMed]
- Foukakis T, Lövrot J, Matikas A, et al. Immune gene expression and response to chemotherapy in advanced breast cancer. Br J Cancer 2018;118:480-8. [Crossref] [PubMed]
- De La Cruz LM, McDonald ES, Mick R, et al. Anti-HER2 CD4(+) T-Helper Type 1 Immune Response is Superior to Breast MRI for Assessing Response to Neoadjuvant Therapy in Patients with HER2-Positive Breast Cancer. Ann Surg Oncol 2017;24:1057-63. [Crossref] [PubMed]
- Cimino-Mathews A, Thompson E, Taube JM, et al. PD-L1 (B7-H1) expression and the immune tumor microenvironment in primary and metastatic breast carcinomas. Hum Pathol 2016;47:52-63. [Crossref] [PubMed]
- Emens LA. Breast cancer immunobiology driving immunotherapy: vaccines and immune checkpoint blockade. Expert Rev Anticancer Ther 2012;12:1597-611. [Crossref] [PubMed]
- Egelston CA, Avalos C, Tu TY, et al. Human breast tumor-infiltrating CD8(+) T cells retain polyfunctionality despite PD-1 expression. Nat Commun 2018;9:4297. [Crossref] [PubMed]
- Thu CA, Chen WV, Rubinstein R, et al. Single-cell identity generated by combinatorial homophilic interactions between alpha, beta, and gamma protocadherins. Cell 2014;158:1045-59. [Crossref] [PubMed]
- Garrett AM, Weiner JA. Control of CNS synapse development by {gamma}-protocadherin-mediated astrocyte-neuron contact. J Neurosci 2009;29:11723-31. [Crossref] [PubMed]
- Frank M, Ebert M, Shan W, et al. Differential expression of individual gamma-protocadherins during mouse brain development. Mol Cell Neurosci 2005;29:603-16. [Crossref] [PubMed]
- Ping Z, Siegal GP, Harada S, et al. ERBB2 mutation is associated with a worse prognosis in patients with CDH1 altered invasive lobular cancer of the breast. Oncotarget 2016;7:80655-63. [Crossref] [PubMed]
- Song J, Wu S, Xia X, et al. Cell adhesion-related gene somatic mutations are enriched in aggressive papillary thyroid microcarcinomas. J Transl Med 2018;16:269. [Crossref] [PubMed]
- Willis SN, Tellier J, Liao Y, et al. Environmental sensing by mature B cells is controlled by the transcription factors PU.1 and SpiB. Nat Commun 2017;8:1426. [Crossref] [PubMed]
- Takagi Y, Shimada K, Shimada S, et al. SPIB is a novel prognostic factor in diffuse large B-cell lymphoma that mediates apoptosis via the PI3K-AKT pathway. Cancer Sci 2016;107:1270-80. [Crossref] [PubMed]
- Yang Y, Shaffer AL 3rd, Emre NC, et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell 2012;21:723-37. [Crossref] [PubMed]
- Sokalski KM, Li SK, Welch I, et al. Deletion of genes encoding PU.1 and Spi-B in B cells impairs differentiation and induces pre-B cell acute lymphoblastic leukemia. Blood 2011;118:2801-8. [Crossref] [PubMed]
- Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature 2007;446:758-64. [Crossref] [PubMed]
- Ho YJ, Lin YM, Huang YC, et al. Tissue microarray-based study of hepatocellular carcinoma validating SPIB as potential clinical prognostic marker. Acta Histochem 2016;118:38-45. [Crossref] [PubMed]
- Rappold I, Ziegler BL, Kohler I, et al. Functional and phenotypic characterization of cord blood and bone marrow subsets expressing FLT3 (CD135) receptor tyrosine kinase. Blood 1997;90:111-25. [PubMed]
- Fischer M, Schnetzke U, Spies-Weisshart B, et al. Impact of FLT3-ITD diversity on response to induction chemotherapy in patients with acute myeloid leukemia. Haematologica 2017;102:e129-31. [Crossref] [PubMed]
- Hoehn D, Medeiros LJ, Chen SS, et al. CD117 expression is a sensitive but nonspecific predictor of FLT3 mutation in T acute lymphoblastic leukemia and T/myeloid acute leukemia. Am J Clin Pathol 2012;137:213-9. [Crossref] [PubMed]
- Jiao X, Wood LD, Lindman M, et al. Somatic mutations in the Notch, NF-KB, PIK3CA, and Hedgehog pathways in human breast cancers. Genes Chromosomes Cancer 2012;51:480-9. [Crossref] [PubMed]
- Ling J, Kumar R. Crosstalk between NFkB and glucocorticoid signaling: a potential target of breast cancer therapy. Cancer Lett 2012;322:119-26. [Crossref] [PubMed]
- Tuveson D, Rai KR. Augmenting NF-kappaB in poor-risk CLL: A general paradigm for other cancers? J Exp Med 2015;212:830-1. [Crossref] [PubMed]
- Shain AH, Garrido M, Botton T, et al. Exome sequencing of desmoplastic melanoma identifies recurrent NFKBIE promoter mutations and diverse activating mutations in the MAPK pathway. Nat Genet 2015;47:1194-9. [Crossref] [PubMed]
- Imamura H, Yoshina S, Ikari K, et al. Impaired NFKBIE gene function decreases cellular uptake of methotrexate by down-regulating SLC19A1 expression in a human rheumatoid arthritis cell line. Mod Rheumatol 2016;26:507-16. [Crossref] [PubMed]
- Tawfik O, Kimler BF, Karnik T, et al. Clinicopathological correlation of PD-L1 expression in primary and metastatic breast cancer and infiltrating immune cells. Hum Pathol 2018;80:170-8. [Crossref] [PubMed]
- Bense RD, Sotiriou C, Piccart-Gebhart MJ, et al. Relevance of Tumor-Infiltrating Immune Cell Composition and Functionality for Disease Outcome in Breast Cancer. J Natl Cancer Inst 2016. [Crossref] [PubMed]


