
LncRNA MATN1-AS1 prevents glioblastoma cell from proliferation and invasion via RELA regulation and MAPK signaling pathway
Introduction
Glioblastoma (GBM) is a type of aggressive brain tumor which exhibited dismal outcome. Median overall survival (OS) of GBM is just 14.6 months. Most of the patients receiving standard treatments including maximal surgical debulking, adjuvant radiotherapy and oral temozolomide would die within two years (1,2). Identification of sensitive and specific GBM targets and development of novel therapeutic strategies has long been an urgent need in clinical practice. However, the molecular mechanism of GBM’s pathogenesis is still elusive. Recent studies have suggested long non-coding RNAs (lncRNAs) as tumor inhibitors or oncogenes, resulting in glioma initiation and progression. This may implicate lncRNAs as potential clinical therapeutic targets in the treatment of GBM (3).
LncRNAs are more than 200 nucleotides in length and function as mediators of mRNA stability, splicing, and recruitment of transcription factors (4,5). LncRNAs have been exhibited an inhibitory role in some malignant cells in vivo and in vitro, such as GAS5 in prostate cancer and brain-derived neurotrophic factor antisense in esophageal cancer (6). Some lncRNAs potentially involved in the development of glioma have been discovered, such as metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), Prader Willi/Angelman region RNA 5, and cancer susceptibility candidate 2 (CASC2) (7-9). Therefore, this study first analyzed the expression profiles of brain glioma and identified MATN1-AS1 as the most significantly downregulated lncRNA using GEO DataSets (GSE15824) analysis. Furthermore, lncRNA MATN1-AS1 was found to directly interact with the RELA gene. RELA, a member of the Rel proteins family (p65, p50, p52, c-Rel, and RelB), was known to interact with the inhibitor of κB (IκB) proteins in the cytoplasm (10). Existing evidence has shown that decreased A20 expression prevents the growth and survival of GBM stem cells by reducing RELA phosphorylation and inhibiting cell cycle progression (11). More importantly, recent evidence has shown that increased MAPK signaling is related to the progression of anaplastic astrocytoma to malignant gliomas (12). Thus, the current study hypothesized that the lncRNA MATN1-AS1 contributes to GBM development by RELA gene regulation and interactions with the MAPK signaling pathway.
Methods
Cells and tissue preparation
Human GBM cells U87MG and U251 (ATCC; Manassas, VA, USA) and HEK-293T cells were cultured with Dulbecco’s Modified Eagle Medium (DMEM) medium containing10% fetal calf serum (FCS) and 50 mg/mL penicillin/streptomycin. Fresh GBM tissue specimens were acquired from 75 recently diagnosed GBM patients who underwent surgery between June 1, 2013 and December 30, 2016 at Tongji Hospital (Wuhan, China). Control tissues were acquired from the brain of ten patients suffered from accidental traumatic brain injury. All specimens were immediately snap frozen and stored at −80 °C after surgery before further use. Informed consents were obtained from all the participating patients. This study was approved by the Ethics Committee of Tongji Hospital.
Bio-informatics prediction
The brain glioma-related microarray (GSE15824) expression data and comment probe file were downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo). Gene expression profiles were obtained through the Affymetrix Human Genome U133 Plus 2.0 Array. Gene expression data were processed for background correction and normalization with Affy installation package of R software (13). Non-specific filtration of expression data was conducted using a combination of the linear model from the Limma installation package and Bayesian Statistics with t-tests in order to screen for differently expressed lncRNAs (14). Differently expressed lncRNAs were predicted with Multi Experiment Matrix (MEM, http://biit.cs.ut.ee/mem/). WebGestalt database (http://www.webgestalt.org) was applied for KEGG enrichment analysis of genes to determine the major biochemical metabolic pathways that the genes contributed to (15).
Cell transfection and grouping
U87 and U251 GBM cells were transfected with MATN1-AS1 and/or RELA over-expression plasmids, or small interfering RNAs (siRNAs) targeting MATN1-AS1 or RELA. Over-expression plasmids of MATN1-AS1 or RELA were constructed by introducing BamHI-EcoRI fragments containing MATN1-AS1 or RELA cDNA into the same site in pcDNA3.1. siRNAs targeting MATN1-AS1 or RELA were purchased from GenePharma. The sequences of MATN1-AS1 and RELA siRNA are shown in Table S1. GBM cells were seeded in 6-well plates at a density of 3×105 cells per well and then transfected with plasmids (50 nM) through Lipofectamine 2000 (Invitrogen Inc., Carlsbad, CA, USA). The cells were collected 48 h later and transfection efficiency was measured by RT-qPCR. Stable cell lines were established by selection with Geneticin (G418; Invitrogen, CA, USA).
Full table
RT-qPCR
Total RNA was isolated from cells using TRIzol (15596-026, Invitrogen, Gaithersburg, MD, USA) followed by reverse transcription to cDNA in accordance with the kit instructions (K1621, Fermentas, Maryland, NY, USA). The primer sequences of lncRNA MATN1-AS1, RELA, extracellular signal-regulated kinase (ERK) 1, Bcl-2-associated X protein (Bax), B-cell leukemia-lymphoma-2 (Bcl-2), survivin, matrix metalloproteinase-9 (MMP-9), and β-actin were produced by Shanghai Genechem Co., Ltd. (Shanghai, China) and are summarized in Table 1. RT-qPCR (Takara Biotechnology Ltd., Dalian, China) was performed using ABI 7500 (ABI, Foster City, CA, USA) to determine mRNA expression of the target genes.

Full table
Immunoblotting analysis
Total proteins were extracted with RIPA buffer and separated on 10% SDS-PAGE. Proteins were then transferred onto polyvinylidene fluoride (PVDF) membranes (HVLP04700, Millipore, Bedford, MA, USA), blocked by 5% skimmed milk at room temperature for 1 h, and incubated at 4 °C overnight with primary antibodies for rat anti-RELA (1:1,000, ab76228), ERK1/2 (1:1,000, ab17942), p-ERK1/2 (1:500, ab223500), Bcl-2 (1:1,000, ab32124), Bax (1:1,000, ab32503), survivin (1:5,000, ab76424), MMP-9 (1:1,000, ab38898), and rabbit anti β-actin (ab8227, Abcam Inc., Cambridge, UK). Horseradish peroxidase (HRP)-labeled goat anti-rabbit IgG served as secondary antibody (1:2,000, ab6721, Abcam Inc., MA, USA) at room temperature for 2 h. Images were taken using the Gel imaging analysis system (Gel Doc XR, Bio-Rad, Hercules, CA, USA). Relative protein expression was defined as the ratio of target protein to the internal reference protein.
MTT methods
GBM cell proliferation was detected by MTT assay and cell counting methods. Briefly, cells were harvested and seeded into a 96-well plate with a density of 1×104 cells/well after 48 h of transfection. On the next day, 10 µL of 5 mg/mL MTT (ST316, Beyotime Biotechnology Co., Shanghai, China) solution was added into each well and cells were incubated at 37 °C for 4 h. Then, DMEM was replaced by 100 µL DMSO for 10 min of incubation at room temperature. The absorbance was measured at 490 nm wavelength.
Transwell invasion assay
After 48 h of transfection, the cells (1×105 cells/well) were seeded into the upper well of a Transwell chamber coated with Matrigel and cultured in DMEM without serum. The lower chamber was full of DMEM including 10% FBS. The cells were incubated at 37 °C for 24 h. Cells on top of the apical chamber were wiped out with a wet cotton swab, fixed with 4% paraformaldehyde for 15 min, and stained with 0.5% crystal violet solution for 30 min. Five fields were randomly selected and photographed under a light microscope.
Flow cytometry
After transfecting for 48 h, the cells were trypsinized, pelleted, and then incubated with pre-cooled 70% ethanol at 4 °C overnight. On the following day, 10 µL of RNase was added and cells were cultivated at 37 °C for 5 min. The cells were then stained with 1% propidium iodide (PI) for 30 min in the dark. Then, cell cycle was analyzed via flow cytometer (FACS Cali-bur, BD, FL, NJ, USA) at an excitation wavelength of 488 nm.
Apoptosis assay
After transfecting for 48 h, cells were digested using EDTA-free pancreatin and centrifuged at 1,000 rpm for 5 min at 4 °C. Cell apoptosis was determined with annexin V-fluorescein isothiocyanate (FITC)/PI double-staining kit (BD Bioscience) following the manufacturer’s instructions. Briefly, a combined mixture of annexin V-FITC and binding buffer (1:40) was used to resuspend the cells. Cells were incubated for 30 min at room temperature. PI and binding buffer mixture (1:40) were then added for 15 min at room temperature. Apoptosis was detected via flow cytometry.
In vivo xenograft tumor growth
Human GBM xenografts were generated via injecting 5×106 of parent or MATN1-AS1 over-expressing U87 cells subcutaneously into the right hind limb of BALB/c athymic nude mice of 6–8 weeks old purchased from Shanghai SLAC Laboratory Animal Co., Ltd. Tumor size was assessed every three days via calipers (volume = length × width × width ×0.5). After 24 days, mice were sacrificed and the tumor tissues were weighed. Animal experiments were approved by Institutional Animal Care Committee and conducted in accordance to the institutional and university guidelines on the care and use of experimental animals.
Immunohistochemistry (IHC)
For mouse xenografts, the primary tumors were fixed with 10% formaldehyde, embedded with paraffin, and then sectioned into serial slices with a thickness of 4 µm. IHC was conducted to analyze RELA and Ki-67 expression based on the manufacturer’s instructions. In brief, after incubation at 4 °C overnight with primary rat anti-human antibodies RELA (1:100, ab16363, Abcam, Cambridge, MA, USA) and Ki67 (1:50, ab8191, Abcam, Cambridge, MA, USA), samples were incubated with HRP-labeled goat anti-rat secondary antibody (ab205718, Abcam, Cambridge, MA, USA). Negative controls (NC) were acquired by eliminating the primary antibody. Images were obtained and analyzed based on at least five random fields of 3 to 5 slides from different mice.
RNA pull-down assay
To investigate the interaction between MATN1-AS1 and E2F6 protein in U87 cells, we conducted RNA pull-down assay according to a Pierce™ Magnetic RNA-Protein Pull-Down Kit (20164, Pierce, Thermo Fisher Scientific Inc., Waltham, Massachusetts, USA) following the manufacturer’s instructions. Biotin-labeled MATN1-AS1 (1 µg) was mixed with structure buffer to obtain RNA with a suitable second-level structure, heated at 95 °C for 2 min, incubated on ice for 3 min, then placed at room temperature for 30 min. The beads were resuspended with 50 µL wash buffer, added to the biotin-labeled denatured RNA, incubated at 4 °C overnight, and centrifuged at 3,000 rpm for 60 s. The magnetic beads/RNA mixture was mixed with U87 cell lysate containing a suitable amount of RNase inhibitor. Then the incubated magnetic beads/RNA-protein mixture was centrifuged with a low speed and the supernatant was recovered. RNA protein complexes were obtained at room temperature with 25 µL streptavidin agarose beads (RITITRONT) for 1 h. The samples were mixed with 5× sodium dodecyl sulphate (SDS) loading buffer and then denatured at 95 °C for 10 min. E2F6 protein expression was detected by immunoblotting.
RNA-binding protein immunoprecipitation (RIP)
RIP was conducted using an EZMagna RIP kit (Shanghai Haoran Biological Technology Co., Ltd., Shanghai, China). In brief, the U87 cell suspension was prepared using equal volume of RIP lysis buffer (P0013B, Beyotime, Shanghai, China). In each co-precipitation reaction system, 75 µL of magnetic bead was resuspended in 100 µL of RIP washing buffer and mixed with 3 µg E2F6 protein antibody (ab11952, Abcam Inc., Cambridge, MA, USA) and 3 µg IgG antibody (ab205718, Abcam Inc., Cambridge, MA, USA) as an NC. The complex of magnetic beads/antibody was washed and then resuspended using 900 µL RIP washing buffer, and incubated for 6 h at 4 °C with 100 µL cell extraction. The sample was put on the magnetic base to collect the complex of magnetic bead/protein using RIP wash buffer. RNA was extracted following 30 min incubation with 0.1% standard deviations and 0.5 mg/mL proteinase K at 55 °C.
Chromatin immunoprecipitation (CHIP) assay
To investigate E2F6 and RELA binding, the CHIP assay was conducted using a CHIP kit (P2078, Beyotime Biotechnology Co., Shanghai, China). Briefly, U87 cells were fixed with a mixture of 37% formaldehyde solution and the culture medium. Crosslink was carried out in an incubator for 10 min at 37 °C. The cells were washed twice with pre-cooled PBS solution by centrifugation at 2,000 rpm 4 °C for 5 min. Then, 1 mL SDS lysis buffer containing PMSF and proteinase inhibitor cocktail was added and kept on ice for 10 min. DNA fragments ranging from 200 to 1,000 bp were obtained by sonication. The fragments in the supernatant were collected by decentering the supernatant at 13,000 rpm for 10 min at 4 °C. Protein A/salmon sperm DNA agar (75 µL) was then added into the supernatant and then incubated on ice for 30 min. The agar was precipitated by decentering at 1,000 rpm for 1 min at 4 °C. The supernatant was transferred to a new EP tube, and 10 µL supernatant was stored in a refrigerator at −20 °C as input control. The E2F6 antibody (3.0 µg, sc-22823, Santa Cruz, Dallas, TX, USA) was added to the supernatant and shaken overnight in a refrigerator at 4 °C. The same amount of mouse IgG antibody (3.0 µg, C15410206, Diagenode, Seraing, Belgium) was used as control. The combined agarose beads with 60 µL protein A/salmon essence DNA were shaken on ice for 60 min, then centrifuged at 1,000 rpm for 1 min at 4 °C to precipitate the agarose beads. Beads were then washed with 1 mL of each washing buffer, gently shaken at room temperature for 5 min, and centrifuged at 1,000 rpm for 1 min at 4 °C. In order to dissociate the E2F6/RELA promoter complex and GATA6 antibody, 100 µL elution buffer was added to the sample, mixed well, and incubated for 15 min at room temperature, and then centrifugated at 1,000 rpm for 1 min. The supernatant was transferred to a new EP tube and eluted with 200 mL buffer. The input was removed and 200 mL elution buffer was added as positive control. Cross-linking was liberated by adding 20 µL of 5 M NaCl. Samples were sequentially incubated overnight in an incubator at 65 °C. The supernatant was added to 1 µL proteinase K and incubated at 37 °C for 1 h. The purified DNA was used as a template DNA for PCR reaction. The E2F6 and RELA promoter binding sites were forward: 5'-CCATCTGGAGAAGACTCCGC-3' and reverse: 5'-AATCTAGATGCGGCTGTGGG-3'.
Dual luciferase reporter assay
RELA promoter region was cloned into luciferase construct pGL3-Basic Vector (Promega, Madison, WI, USA) to produce pGL3-RELA promoter-luc. Then, the wild-type (wt) and mutated (mut) sequences of the binding site of RELA promoter region and E2F6 were sub-cloned into the pGL3-RELA promoter-luc. After that, the RELA mut and RELA wt plasmids were co-transfected with E2F6 or NC into HEK-293T cells (AT-1592, ATCC, Manassas, VA, USA). Renilla luciferase was used as the internal control. Finally, 48 h after transfection, firefly and Renilla luciferase were detected using a luciferase assay kit (K801-200, Biovision, USA) on a dual-luciferase reporter system (Promega).
Statistical analysis
SPSS 16.0 software (SPSS Inc., Chicago, IL, USA) was used to analyze data. Comparisons between groups were processed using Student’s t-test or one-way analysis of variance (ANOVA). OS curves were calculated by combining Kaplan-Meier method and the log-rank test. Correlations was determined by Pearson correlation coefficient analysis. A P value <0.05 was considered statistically significant.
Results
MATN1-AS1 was downregulated in GBM with poor prognosis
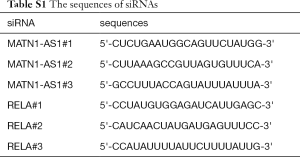
Analysis of GBM-related microarray GSE15824 revealed that MATN1-AS1 expression showed the largest significant decrease (Figure 1A). Furthermore, bioinformatics analysis of gene expression in GBM using The Cancer Genome Atlas (TCGA) indicated that MATN1-AS1 was significantly decreased in GBM compared with normal tissues (P=0.0002, Figure 1B). Next, MEM was used to predict the top 100 co-expression genes associated with MATN1-AS1 and KEGG was used to conduct a pathway enrichment analysis. It was found that only RELA was correlated with the MAPK signaling pathway, which was highly involved in GBM pathogenesis (Figure 1C). The TCGA database further confirmed that MATN1-AS1 was negatively correlated with RELA (r=−0.2398, P=0.0017, Figure 1D). Collectively, MATN1-AS1 was involved in GBM progression.
To confirm these findings, 75 GBM tumors and ten normal brain tissues were collected and mRNA levels of MATN1-AS1 and RELA were quantified using real time RT-PCR. As depicted in Figure 1E, MANTN1-AS1 was markedly downregulated (P=0.001), whereas RELA was significantly enhanced in GBM tumors compared with normal brain tissues (P=0.008). Moreover, MATN1-AS1 expression was negatively correlated with RELA levels in GBM tissues (r=−0.740, P=0.0001, Figure 1F). Kaplan-Meier survival curve analysis revealed that the arbitrary cut-off used for high and low expression of MATN1-AS1 was 0.66 (mean value) and that higher MATNI-AS1 expression predicted better OS in patients (P=0.004, Figure 1G).
MATN1-AS1 suppressed mRNA and protein expression of MAPK signaling pathway-related genes in GBM cells
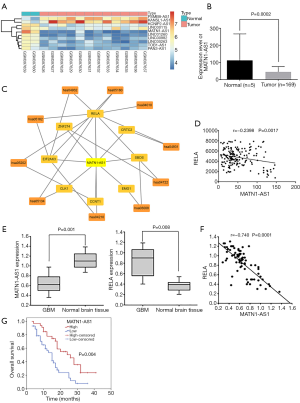
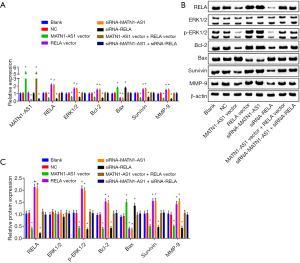
Based on the silencing efficacies of siRNA-MATN1-AS1 and siRNA-RELA (Figure S1A), siRNA-MATN1-AS1#3 and siRNA-RELA#1 were chosen for subsequent experiments. As shown in Figure 2A, RELA, ERK1/2, Bcl-2, survivin, and MMP-9 mRNA expression were obviously decreased in cells with MATN1-AS1 over-expression or siRNA-RELA knockdown. Conversely, they were significantly increased in cells transfected with siRNA-MATN1-AS1 knockdown or RELA over-expression vectors. In contrast, Bax mRNA was significantly decreased in cells with MATN1-AS1 over-expression or siRNA-RELA knockdown, but increased in cells transfected with siRNA-MATN1-AS1 or RELA over-expression vectors. Similar to changes in mRNA expression, MAPK signaling pathway-related protein levels were significantly inhibited or stimulated by each manipulation in the same direction (Figure 2B,C). While MATN1-AS1 expression was diminished in cells transfected with siRNA-MATN1-AS1, it was not significantly changed in cells with RELA over-expression. However, MATN1-AS1 expression was significantly elevated in cells double-transfected with over-expressing MATN1-AS1 vector and RELA vector, but significantly reduced in cells transfected with both siRNA-MATN1-AS1 and siRNA-RELA. No significant differences were found in the aforementioned gene and protein expression between NC (empty vector) and the non-transfected blank group (all P>0.05).
MATN1-AS1 inhibited GBM cell proliferation and promoted cellular apoptosis by RELA inhibition
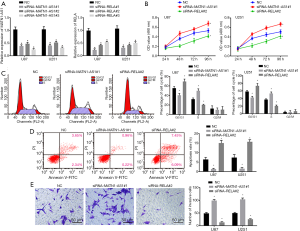
Cell proliferation and survival were examined in GBM cells transfected with siRNA-MATN1-AS1#3, MATN1-AS1 over-expressing vector, siRNA-RELA#1, or RELA over-expressing vector. Cell proliferation was significantly inhibited in cells with MATN1-AS1 over-expression or siRNA-RELA knockdown. Conversely, it was significantly increased in cells with siRNA-MATN1-AS1 knockdown or RELA over-expression (all P<0.05, Figures 3A,S1B). Importantly, compared to NC, cell proliferation was not significantly changed in cells with double-over-expression of MATN1-AS1 and RELA or double-knockdown of MATN1-AS1 and RELA, which indicated that RELA could counteract MATN1-AS1 in regulating cell proliferation. Furthermore, MATN1-AS1 over-expression or siRNA-RELA knockdown resulted in a decrease of S phase proportion in GBM cells, whereas siRNA-MATN1-AS1 knockdown or RELA over-expression led to an increase of S phase fraction (Figures 3B,S1C).
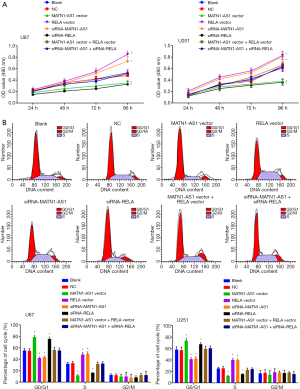
Similarly, cellular apoptosis rates were significantly increased in cells with MATN1-AS1 over-expression or siRNA-RELA knockdown, but lower in cells with siRNA-MATN1-AS1 knockdown cells or RELA over-expression cells (Ps<0.05, Figures 4A,S1D). No significant differences of cell proliferation and apoptosis were noted between NC and non-transfected blank groups, the MATN1-AS1 over-expression and the siRNA-RELA groups, the siRNA-MATN1-AS1 and RELA over-expression groups, or double-over-expression of MATN1-AS1 and RELA compared to double knockdown of MATN1-AS1 and RELA groups (Ps>0.05).
MATN1-AS1 inhibited cell invasion
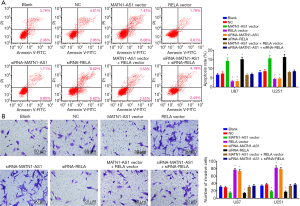
Compared with the NC and blank groups, cell invasion was reduced by MATN1-AS1 over-expression or siRNA-RELA knockdown, but increased by siRNA-MATN1-AS1 knockdown or RELA over-expression (Ps>0.05). There were no significant differences in regard as cell invasion between the MATN1-AS1 over-expression and siRNA-RELA knockdown groups, the RELA over-expression and siRNA-MATN1-AS1 knockdown groups, or the double-over-expression of MATN1-AS1 and RELA and the double knockdown of MATN1-AS1 and RELA group (Ps>0.05) (Figures 4B,S1E).
MATN1-AS1 suppressed tumor growth in vivo
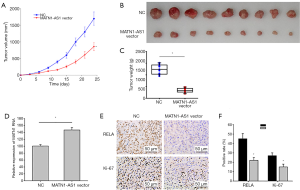
MATN1-AS1 over-expressing cells were injected into nude mice to further investigate the possible role of MATN1-AS1 in vivo. MATN1-AS1 over-expression markedly decreased tumor volume and weight compared to the NCs (Figure 5A,B,C). Further IHC staining of RELA and Ki-67 within tumor xenografts revealed that MATN1-AS1 over-expression (Figure 5D) resulted in inhibition of GBM cell proliferation (Figure 5E,F) and downregulated RELA expression (Figure 5E,F).
MATN1-AS1 inhibited the expression of RELA
RPISeq analysis (http://pridb.gdcb.iastate.edu/RPISeq/) revealed that MATN1-AS1 bound to E2F6 transcription factor. Therefore, the interaction between MATN1-AS1 and E2F6 was investigated. The RNA pull-down assay showed that MATN1-AS1 could bind to E2F6 protein (Figure 6A), and the RIP assay showed that MATN1-AS1 binding to E2F6 was significantly increased compared to IgG antibody binding (Figure 6B).
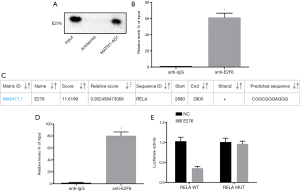
A search of the JASPAR database (http://jaspar.genereg.net/) revealed that E2F6 transcription factor could bind to RELA promotor (Figure 6C). CHIP analysis showed that a large number of promoter fragments of the RELA gene could be detected in DNA samples recovered by E2F6 antibody immunoprecipitation, suggesting that E2F6 could bind to the RELA promoter region (Figure 6D). The dual luciferase reporter gene assay further demonstrated that E2F6 bound to the RELA promoter region (Figure 6E). Luciferase activity in the wild-type RELA and E2F6 co-transfection group was markedly decreased compared to the NC group (P<0.05), whereas it was not different between the mutant type RELA and E2F6 co-transfection and NC groups (P>0.05).
Discussion
GBM is an extremely progressive brain tumor (16). Despite intensive treatment with conventional therapeutic protocols, majority of the GBM patients succumb within two years. It is well-known that lncRNAs are powerful transcriptional and post-transcriptional molecules that regulate gene activity and play key roles in cancer development (17). Therefore, we attempted to explore the role of lncRNA MATN1-AS1 in brain GBM.
Increasing evidence has demonstrated that lncRNAs play a part in the regulation of gene expression via chromatin modification, transcription, post-transcription and other processes (18). For example, previous work has shown that lncRNA CASC2 expression was decreased in glioma tissues and cell lines and related to shorter survival time of patients with glioma (19). Further, over-expression of the lncRNA ADAMTS9-AS2, caused by knocking down DNA methyltransferase-1, resulted in significant cell migration inhibition and cell apoptosis enhancement in glioma, whereas IncRNA ADAMTS9-AS2 knockdown resulted in the opposite effects (20). The current study first reports that lncRNA MATN1-AS1 serves as a tumor suppressor in the development of GBM. MATN1-AS1 expression was significantly downregulated in GBM tissues compared to normal brain tissues. Lower expression of MATN1-AS1 predicted poorer outcomes for patients with GBM. siRNA-MATN1-AS1 knockdown resulted in increased mRNA and protein Bcl-2 levels, but reduced Bax expression. Furthermore, MATN1-AS1 expression was negatively correlated with RELA in GBM, and the current experiments confirmed that RELA was targeted by MATN1-AS1. MATN1-AS1 over-expression resulted in downregulation of RELA, ERK1/2, Bcl-2, survivin, and MMP-9, but upregulation of Bax. The above results suggest MATN1-AS1 could promote GBM cell apoptosis through regulation of RELA and the apoptotic pathway.
RELA, also known as p65, is a member in NF-κB family. Abnormal RELA expression is linked to improper development, inflammatory diseases, and cancer (21). Studies have revealed that spinal NF-κB/RELA promotes the development of peripheral inflammation and hyperalgesia (22). Acupuncture was found to relieve pain after spinal cord injury by inhibiting reactive oxygen species-induced p38MAPK and ERK activation (23). Que has also been shown to function as a spinal cord protector via inhibition of the p38MAPK/iNOS signaling pathway (24). As a member of the apoptosis inhibitor protein family, survivin—an inhibitor of caspases and cell death—was found to be over-expressed in many cancers and is often related to poor clinical outcomes (25). MMP-9 may play a role in many brain disorders, such as schizophrenia, epilepsy, autism spectrum disorder, stroke, brain injury, and brain tumors (26). The current results consistently demonstrated that siRNA-MATN1-AS1 knockdown resulted in significant increases in Bcl-2 and survivin, inhibition of Bax, promoted GBM cell apoptosis, and inhibited GBM cell invasion. Although apoptosis may result in less cancer cell migration, the current study found that the number of migrated cells in the trans-wells was markedly reduced in the siRNA-MATN1-AS1 cells even after correcting for the apoptosis effect. This suggests that siRNA-MATN1-AS1 knockdown not only promotes apoptosis, but also inhibited cell migration.
Most importantly, through inhibiting the components of the MAPK signaling pathway, MATN1-AS1 inhibited GBM cell proliferation and invasion, but promoted GBM cell apoptosis. Previous work has shown that lncRNAs can regulate critical cell cycle regulators (cyclin-dependent kinases and inhibitors, cyclins, p53) and participated in cell cycle arrest or apoptosis in response to DNA damage (27). In stomach cancer, downregulation of the lncRNA GAS5 can abolish G1 phase cell cycle arrest through inhibition of Y-box binding protein 1 expression (28). Signaling molecules in the MAPK cascade can relay, amplify, and integrate various signaling pathways, thereby allowing a large variety of cellular processes, including cell proliferation. Blocking the MAPK signaling pathway has also been shown to inhibit cell invasion in GBM cells (29). Furthermore, it has been shown that MAPK signaling pathway activation can increase cell survival through the post-translational modification and inactivation of cell death components, as well as the upregulation of pro-survival gene transcription (30).
In conclusion, the current study demonstrates that lncRNA MATN1-AS1 upregulation suppressed cell proliferation and invasion, but accelerated apoptosis of GBM cells through downregulation of RELA and inhibition of the MAPK signaling pathway, suggesting that MATN1-AS1 could be a potential therapeutic target for GBM.
Acknowledgments
Funding: This work was funded by Program of the National Natural Science Foundation of China (grant number: 81772680).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Ethical approval was given by the Ethics Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology. Informed consent was obtained from all patients. Animal experiments were approved by Institutional Animal Care Committee and conducted in accordance to the institutional and university guidelines on the care and use of experimental animals.
References
- Kim BS, Seol HJ, Nam DH, et al. Concurrent Chemoradiotherapy with Temozolomide Followed by Adjuvant Temozolomide for Newly Diagnosed Glioblastoma Patients: A Retrospective Multicenter Observation Study in Korea. Cancer Res Treat 2017;49:193. [Crossref] [PubMed]
- Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352:987-96. [Crossref] [PubMed]
- Zhang XQ, Leung KK. Long non-coding RNAs in glioma: Functional roles and clinical perspectives. Neurochem Int 2014;77:78-85. [Crossref] [PubMed]
- Bergmann JH, Spector DL. Long non-coding RNAs: modulators of nuclear structure and function. Curr Opin Cell Biol 2014;26:10-8. [Crossref] [PubMed]
- Dey BK, Mueller AC, Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription 2014;5:e944014. [Crossref] [PubMed]
- Li CH, Chen Y. Targeting long non-coding RNAs in cancers: Progress and prospects. Int J Biochem Cell Biol 2013;45:1895-910. [Crossref] [PubMed]
- Park JY, Lee JE, Park JB, et al. Roles of Long Non-Coding RNAs on Tumorigenesis and Glioma Development. Brain Tumor Res Treat 2014;2:1-6. [Crossref] [PubMed]
- Wang P, Liu YH, Yao YL, et al. Long non-coding RNA CASC2 suppresses malignancy in human gliomas by miR-21 ☆. Cell Signal 2015;27:275-82. [Crossref] [PubMed]
- Wang XP, Shan C, Deng XL, et al. Long non-coding RNA PAR5 inhibits the proliferation and progression of glioma through interaction with EZH2. Oncol Rep 2017;38:3177. [Crossref] [PubMed]
- Yang N, Wang P, Wang WJ, et al. Inhibition of cathepsin L sensitizes human glioma cells to ionizing radiation in vitro through NF-κB signaling pathway. Acta Pharmacol Sin 2015;36:400. [Crossref] [PubMed]
- Hjelmeland AB, Wu Q, Wickman S, et al. Targeting A20 decreases glioma stem cell survival and tumor growth. PLoS Biol 2010;8:e1000319. [Crossref] [PubMed]
- Mizoguchi M, Betensky RA, Batchelor TT, et al. Activation of STAT3, MAPK, and AKT in malignant astrocytic gliomas: correlation with EGFR status, tumor grade, and survival. J Neuropathol Exp Neurol 2006;65:1181-8. [Crossref] [PubMed]
- Fujita A, Sato JR, Rodrigues Lde O, et al. Evaluating different methods of microarray data normalization. BMC Bioinformatics 2006;7:469. [Crossref] [PubMed]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004;3:Article3.
- Wang J, Duncan D, Shi Z, et al. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res 2013;41:W77-83. [Crossref] [PubMed]
- Zinn PO, Colen RR, Kasper EM, et al. Extent of resection and radiotherapy in GBM: A 1973 to 2007 surveillance, epidemiology and end results analysis of 21,783 patients. Int J Oncol 2013;42:929-34. [Crossref] [PubMed]
- Yang X, Song JH, Cheng Y, et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut 2014;63:881. [Crossref] [PubMed]
- Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464:1071-6. [Crossref] [PubMed]
- Liao Y, Shen L, Zhao H, et al. LncRNA CASC2 Interacts With miR-181a to Modulate Glioma Growth and Resistance to TMZ Through PTEN Pathway. J Cell Biochem 2017;118:1889-99. [Crossref] [PubMed]
- Yao J, Zhou B, Zhang J, et al. A new tumor suppressor LncRNA ADAMTS9-AS2 is regulated by DNMT1 and inhibits migration of glioma cells. Tumour Biol 2014;35:7935-44. [Crossref] [PubMed]
- Kim MY, Koh DI, Choi WI, et al. ZBTB2 increases PDK4 expression by transcriptional repression of RelA/p65. Nucleic Acids Res 2015;43:1609. [Crossref] [PubMed]
- Luo JG, Zhao XL, Xu WC, et al. Activation of Spinal NF-κB/p65 Contributes to Peripheral Inflammation and Hyperalgesia in Rat Adjuvant-Induced Arthritis. Arthritis Rheumatol 2014;66:896-906. [Crossref] [PubMed]
- Choi DC, Lee JY, Lim EJ, et al. Inhibition of ROS-induced p38MAPK and ERK activation in microglia by acupuncture relieves neuropathic pain after spinal cord injury in rats. Exp Neurol 2012;236:268-82. [Crossref] [PubMed]
- Song Y, Liu J, Zhang F, et al. Antioxidant effect of quercetin against acute spinal cord injury in rats and its correlation with the p38MAPK/iNOS signaling pathway. Life Sci 2013;92:1215-21. [Crossref] [PubMed]
- Jaiswal PK, Goel A, Mittal RD. Survivin: A molecular biomarker in cancer. Indian J Med Res 2015;141:389-97. [Crossref] [PubMed]
- Vafadari B, Salamian A, Kaczmarek L. MMP-9 in Translation: From Molecule to Brain Physiology, Pathology and Therapy. J Neurochem 2016;139:91-114. [Crossref] [PubMed]
- Kitagawa M, Kitagawa K, Kotake Y, et al. Cell cycle regulation by long non-coding RNAs. Cell Mol Life Sci 2013;70:4785-94. [Crossref] [PubMed]
- Liu Y, Jing Z, Zhang W, et al. lncRNA GAS5 enhances G1 cell cycle arrest via binding to YBX1 to regulate p21 expression in stomach cancer. Sci Rep 2015;5:10159. [Crossref] [PubMed]
- Krueger JS, Keshamouni VG, Atanaskova N, et al. Temporal and quantitative regulation of mitogen-activated protein kinase (MAPK) modulates cell motility and invasion. Oncogene 2001;20:4209-18. [Crossref] [PubMed]
- Bonni A, Brunet A, West AE, et al. Cell Survival Promoted by the Ras-MAPK Signaling Pathway by Transcription- Dependent and -Independent Mechanisms. Science 1999;286:1358-62. [Crossref] [PubMed]


