
This article has an erratum available at: http://dx.doi.org/10.21037/atm-2022-48 the article has been update on 2022-08-08 at here.
Epigenetic hypomethylation and upregulation of GD3s in triple negative breast cancer
Introduction
Breast cancer has become the second most common cause of mortality and morbidity in women all over the world (1). Breast cancers have many subtypes, but there are three main ones: ER+, HER2+ and triple-negative breast cancer (TNBC) (2). TNBC is a very aggressive cell type with a poor prognosis. Understanding the molecular mechanism of TNBC malignancy should allow researchers to identify new effective drug targets.
Gangliosides are very important in the development (3), differentiation (4,5) and proliferation (6) of cells. Ganglioside D3 (GD3) and ganglioside D2 (GD2) are strongly expressed in neuroectoderm-derived cancers and thought to be important in the malignant transformation of tumor cells (7). The expression profiles of gangliosides in the development and transformation of many cancers have been determined (8,9). GD3 is highly expressed in malignant melanomas (10,11) where it can be used as a biomarker of human melanoma (12) since it is only expressed at low levels in normal melanocytes (13). GD2 is overexpressed in melanomas and small cell lung cancers where it increases the growth and invasiveness of cells (14). GD2 was used as a biomarker of breast stem cells (15) and mesenchymal stem cells (16).
GD3 synthase (GD3s) is one of six members of the ST8Sia subfamily of sialyltransferases (ST8Sia I-VI), also known as ST8SIA1. GD2 and GD3 are highly expressed in many malignant cancers (17-19) and GD3s is the only enzyme that is responsible for biosynthesis of GD2 and GD3 (20). GD3s plays a very important role in the development of cancers (21,22), but the signaling pathways involved in GD3s activity in breast cancers like TNBC have not been fully delineated.
In this study, we explored the expression of GD3s in cells and tissues of different types of breast cancer and found higher GD3s levels in ER− breast cancers than in ER+ breast cancers and a negative correlation with the signature of luminal breast cancer. GD3s overexpression in TNBCs was associated with hypomethylation of the synthase gene, ST8SIA1, and resulted in increased proliferation, invasion, migration and colony formation of cell. Inhibition of GD3s reduced proliferation, invasion, migration and colony formation of cell. Clinically, a lower GD3s expression was clearly associated with relapse-free survival (RFS) and increased overall survival (OS) of breast cancer patients.
Methods
Cell culture
The human breast cancer cell lines, MCF-7, MDA-MB-468 and T47D, were purchased from the American Type Culture Collection. MCF-7 cells were cultured in DMEM, while MDA-MB-468 and T47D were cultured in RPMI 1640. Both mediums were supplemented with 10% fetal calf serum, 100 units/mL penicillin, and 100 µg/mL streptomycin at 37 °C in a humidified incubator containing 5% CO2.
Tumor specimens
The Institutional Review Board at the John Wayne Cancer Institute/Saint John Health Center Providence Health System, Santa Monica, CA, approved the use of human tissues (MORD-RTPCR-0995). Immunohistochemistry (IHC) was carried out on sections from paraffin-embedded archival tissue (PEAT) specimens of breast cancers, which had been diagnosed at the Saint John Health Center Providence Health System, Santa Monica, CA.
Stable transfection
MCF-7 cells were seeded in 60 mm dishes at 70–80% confluence for 24 h before transfection. MCF-7 cells, which do not express GD3s, were transfected with a GD3s expression plasmid using the transfection reagent, Lipofectamine™ 3000, (Invitrogen, Grand Island, NY, USA) for 24 h. The cells were then grown in the presence of 800 µg/mL G418 (Invitrogen, Grand Island, NY, USA) for three weeks. MCF-7 cells with high GD3s expression were referred to as MCF-7-GD3s. MDA-MB-468 cells, which have a normally high GD3s expression level, were plated in 60 mm dishes at 70–80% confluence for 24 h before transfection with GD3s shRNA (Sigma-Aldrich, St. Louis, MO, USA). Stably transfected MDA-MB-468-shRNA cells were selected by incubation with 5 µg/mL puromycin. For controls, MCF-7 cells were transfected with empty vector (MCF-7-cntl) and MDA-MB-468 cells were transfected with a control shRNA (MDA-MB-468cntl).
GD3s expression was verified by Western blotting with anti-GD3s antibody (Abcam, Cat. No. ab214936, Cambridge, UK), anti-myc antibody (EMD Millipore, Cat. No. 06-340, San Diego, CA) and anti-FLAG antibody (Origene, Cat. No. TA50011-100, Rockville, MD, USA).
Cell growth and colony formation in soft agar
Cell proliferation was assessed by MTT [3-(4,5-dimethylthiazol-2-yl)-2,5diphenyltetrazolium] assay (Sigma-Aldrich, St. Louis, MO, USA). The colony formation assay in soft agar was carried out in six-well culture plates. After 2–3 weeks, the colony were stained with 5 mg/mL MTT and the number was counted. Each experiment was done in triplicate.
Cell migration and invasion assay
About 1×104 cells were seeded on top of the Boyden chamber inserts (BD Biosciences, San Jose, CA, USA) and 10% fetal calf serum was used as the chemoattractant. To reduce the effect of cell proliferation, mitomycin C was added to the cells to a final concentration of 2 µg/mL. Cells that had migrated through the membrane onto the lower surface of the inserts were stained with 1% crystal violet and counted in four randomly selected fields using a microscope. For invasion assays, inserts were coated with a thin layer of Matrigel basement membrane matrix. The subsequent steps in the invasion assay were the same as those of the migration assay.
In silico analysis
The Oncomine database (www.oncomine.org) is very useful for investigating genes that are expressed in multiple cancer datasets to validate the relationship between transcription and disease. More advanced analyses were used to check gene expression in a small fraction of samples of a cancer type using different filters. The expression of GD3s mRNA was checked in the subtypes of breast cancers. The Cancer Genome Atlas (TCGA) was initiated by the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI) and can be utilized to study the molecular basis of cancer through the application of genome analysis technologies. The cBioPortal for Cancer Genomics (http://www.cbioportal.org/) provides many different cancer data sets, such as sequencing data, microarray data, RNA-Seq data, etc. The cBioPortal can also be used to assess the effects of co-expression of genes. There is a data set of 1,881 breast tumor samples and a 51-sample breast cancer cell line set available in GOBO (http://co.bmc.lu.se/gobo). Many different analyses can be performed using these data sets, which were all from Affymetrix U133A microarrays. The mRNA expression of specific genes in cancers can be easily checked in GOBO. The association between gene expression and patient outcomes can also be determined by using the GOBO dataset. The website (https://genome-cancer.ucsc.edu/proj/site/hgHeatmap/) was used to assess the methylation of genes in different cancers from TCGA data.
Quantitative reverse transcriptase-PCR (qRT-PCR)
Total RNA (1 µg) was extracted from breast cancer cell lines and used for cDNA synthesis according to the manufacturer’s instructions. (Qiagen, Hilden, Germany). The cDNA was added to PCR mixture that contained 1X SYBR Green PCR master mix (Quanta Biosciences, Gaithersburg, MD) and 300 nmol/L gene-specific GD3s primers (AuGCT). The assays were carried out three times on a CFX thermocycler (Bio-Rad, Hercules, CA). The primers are given in Table 1.

Full table
Quantitative real-time methylation-specific PCR (qMS-PCR)
Genomic DNA was extracted from MCF-7 and T47D breast cancer cells (1×106) using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The bisulfite DNA modification was performed according to the manufacturer’s instructions and the methylation level of the ST8SIA1 gene was determined by qMS-PCR using two primer sets, one designed for methylated (M) DNA and the other for unmethylated (U) DNA. The primers for the methylation-specific PCR and unmethylated-specific PCR of the ST8SIA1 gene (Table 2) were designed using Urogene’s MethPrimer software: (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi).

Full table
IHC
IHC was carried out on five-micron sections from PEAT specimens. Expression of GD3s protein was measured using polyclonal anti-GD3s rabbit antibody (No. HPA026775, Sigma-Aldrich, St. Louis, MO, USA) at a dilution of 1:200. Slides were deparaffinized, then rehydrated and washed in 1X PBS. Antigen retrieval was done with citrate buffer (Sigma-Aldrich) at 100 °C for 10 min. After antigen retrieval, slides were incubated in 3% H2O2 (Sigma-Aldrich) to block endogenous peroxidase at room temperature. The GD3s antibody was added to the slides, which were incubated overnight at 4 °C, followed by 1 h incubation with biotinylated second Ab. Staining was done with diaminobenzidine (Vectastain, Burlingame, CA, USA) and cells were counterstained with hematoxylin (Sigma-Aldrich). The negative controls were treated with rabbit serum alone (Santa Cruz Biotechnology, Dallas, TX, USA). A Nikon Eclipse Ti microscope was used to take photograph of stained sections and staining density was analyzed by Image J software (http://rsbweb.nih.gov/ij/).
Western immunoblotting
Western immunoblotting analysis was done as previously described (23). Cells were lysed in RIPA buffer on ice with protease inhibitors (Applygen Technologies Inc., Beijing, China) and total protein was extracted. Protein lysates were separated on 12% polyacrylamide gels with SDS-Tris-glycine running buffer, transferred to polyvinylidene difluoride (PVDF) membranes, and blocked with 5% milk in TBST. The PVDF membranes were then incubated overnight with primary antibodies for GD3s (Abcam, No. ab37806, Cambridge, UK) and GAPDH (No. 10494-1-AP, Proteintech, Wuhan, China). After washing with TBST, the blots were incubated with HRP-conjugated secondary antibodies (No. 7074, anti-rabbit IgG; No. 7076, anti-mouse IgG, Cell Signaling Technology, Danvers, USA). Signal detection was performed using the Pierce™ ECL Western blotting substrate (Thermo Fisher Scientific, Waltham, MA, USA) and Bio-Rad Imaging System (Hercules, USA).
Statistical analysis
The results are given as mean ± SD. Statistical significance between different group was assessed by Student’s t-test. A P
Results
GD3s was inversely associated with signature biomarkers of luminal breast cancers: ESR1, FOXA1, GATA3, XBP1 and MYB
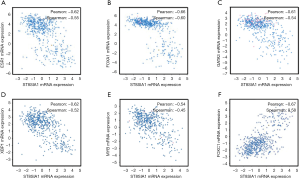
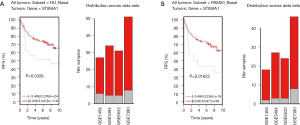
To explore the association between GD3s and the biomarkers of luminal breast cancers, we downloaded and analyzed the co-expression data for GD3s from TCGA (http://www.cbioportal.org/public-portal/cross_cancer.do). The results showed that GD3s mRNA expression was inversely correlated with expression of the luminal breast cancer markers, ESR1, GATA3, FOXA1, MYB and XBP1 mRNA (Figure 1A,B,C,D,E). The expression of GD3s mRNA was positively associated with FOXC1, which is a biomarker of TNBC (24) (Figure 1F). Overall, these results suggest that GD3s may be a potential biomarker of luminal breast cancer and TNBC.
Expression of GD3s in ER- breast cancers was higher than in ER+ breast cancers
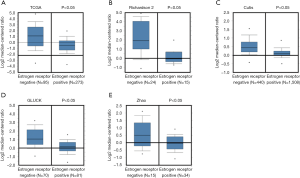
To test the association between GD3s and ER in breast cancers, we did an in silico assay to compare the expression of GD3s in breast cancer patients with ER+ or ER- cell types using the Oncomine database. There was much higher GD3s expression in breast cancer patients that were ER- compared to those who were ER+ (Figure 2).
GD3s was highly expressed in TNBCs
We compared GD3s expression in TNBCs with that in other types of breast cancers using data from Oncomine and TCGA. Expression of GD3s was much higher in TNBCs than other types of breast cancer (Figure 3A,B,C,D,E).
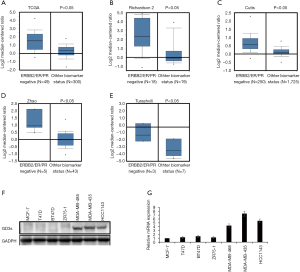
To determine the levels of GD3s mRNA and protein expression in breast cancer cell lines, qRT-PCR and Western blotting were carried out. The expression of GD3s mRNA and protein in TNBC cells was significantly higher than that in other breast cancer cell lines (Figure 3F,G).
GD3s promoted proliferation, invasion, migration and colony formation of breast cancer cells
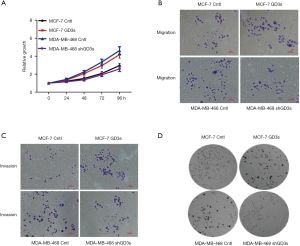
To investigate the functional roles of GD3s in breast cancer, MCF-7 cells which normally have low GD3s expression were transfected with a GD3s expression plasmid. The MDA-MB-468 cell line, which normally has high GD3s expression, was transfected with GD3s shRNA that blocks transcription of the GD3s gene. Controls (cntl) were empty vector for MCF-7 and control shRNA for the MDA cell line. MCF-7-cntl, MCF-7-GD3s, MDA-MB-468-cntl, and MDA-MB-468-shRNA were tested for cell proliferation, migration, invasion and colony formation. Overexpression of GD3s promoted proliferation, invasion, migration and colony formation, whereas knockdown of GD3s inhibited these functions (Figure 4).
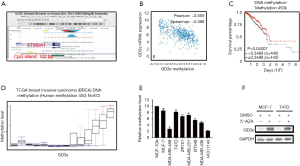
GD3s expression was associated with methylation of the ST8SIA1 gene
To determine if GD3s expression was associated with methylation of the ST8SIA1 gene, in silico analysis using the UCSC gene browser (http://genome.ucsc.edu) was carried out. Results showed that there was a 102 bp CpG island in the promoter region of the ST8SIA1 gene (Figure 5A and Figure S1). Data from TCGA demonstrated that expression of GD3s was inversely correlated with methylation of the ST8SIA1 gene (Figure 5B) and that survival rate of patients with breast cancers was significantly associated with methylation of ST8SIA1 gene (Figure 5C). Results from a methylation array of TCGA data (https://genome-cancer.ucsc.edu/proj/site/hgHeatmap/) showed that there was significantly less methylation of ST8SIA1 gene in TCGA breast cancer tissues (Figure 5D). To further explore the association between GD3s expression and methylation of ST8SIA1 gene in breast cancer cells, MS-PCR was used to assess the methylation level in the ST8SIA1 gene promoter in eight breast cancer cell lines: MCF7, MDA-MB-468, T47D, ZR751, MDA-MB-231, BT549, MDA-MB-436, and HCC1143, plus a non-tumor cell line MCF-10A. The methylation levels of the ST8SIA1 gene promoter in the breast cancer cell lines were significantly lower (PFigure 5E). To further explore if ST8SIA1 gene expression was regulated by methylation, we treated MCF-7 cells and T47D cells, which had low GD3s expression, with the methyl transferase inhibitor, 5-azacytidine, or with vehicle control (DMSO). GD3s expression in MCF-7 cells and T47D cells incubated with 5-azacytidine was increased compared to controls (Figure 5F). In sum, our results demonstrated that GD3s expression in breast cancer cells was regulated by methylation of ST8SIA1.
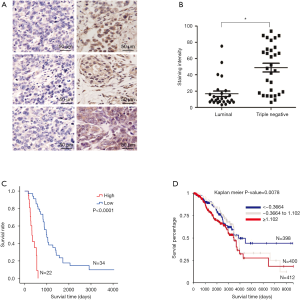
GD3s was overexpressed in TNBC tissues and associated with reduced OS and RFS
IHC was performed to determine if TNBCs had greater GD3s protein levels than estrogen- or progesterone-receptor-positive luminal breast cancers. Results showed that staining density of GD3s was significantly higher in TNBCs compared to luminal breast cancers (Figure 6A,B). To ascertain whether expression of GD3s affected the survival of patients with breast cancer, Kaplan Meier analyses were carried out. It was shown that patients with high GD3s had lower survival rates, while patients with low GD3s had higher survival rates (Figure 6C). Results from an in silico assay confirmed the survival data (Figure 6D) and showed that low GD3s expression was significantly associated with RFS of breast cancer patients (Figure S2).
Discussion
Breast cancers are commonly classified into three subtypes, ER+, HER2+ and TNBC (2). TNBC has the worst prognosis. Up to now, there has been no drug specifically targeting TNBC and the molecular mechanisms controlling TNBC’s malignancy were unclear. Here, we reported that the ganglioside synthase, GD3s, was associated with luminal breast cancer and highly expressed in ER− tumors and TNBC. Expression of GD3s was regulated by methylation of the synthase gene, ST8SIA1 and promoted the proliferation, migration invasion and colony formation of cancer cells. In terms of survival, higher expression levels of GD3s were significantly associated with lower RFS and lower OS of breast cancer patients.
GD2 and GD3 are highly expressed in a variety of malignant tumors (25). GD3s is the sole enzyme responsible for biosynthesis of GD3 and GD2 (20). Recent studies showed that GD3s was highly expressed in many cancers and was very important in the development of cancers (25,26). In this study, data from TCGA showed that GD3s was negatively correlated with the signatures of luminal breast cancers: ESR1, FOXA1, GATA3, XBP1 and MYB. We also found that GD3s expression was much higher in ER- breast cancers than in ER+ and in TNBCs compared to luminal breast cancers. Results from IHC showed that there was much higher expression of GD3s protein in patients with TNBC than other types of breast cancer. All these results suggested that GD3s was not only associated with ER- breast cancers but also with TNBC.
It was reported that cell migration and colony formation in MDA-MB-468 and MDA-MB-231 cell lines was reduced if expression of GD3s was knocked down by GD3s shRNA (27). Our results showed that GD3s over-expression in MCF-7 cells increased proliferation, migration, invasion and colony formation while knockdown of GD3s inhibited proliferation, invasion, migration and colony formation of cells in MDA-MB-468. Our results confirmed previous evidence in part and further suggested that GD3s played an important functional role in the development of breast cancer. Targeting GD3s inhibited proliferation and mobility, suggesting that GD3s may be a feasible drug target for breast cancer therapy, especially for TNBC.
Epigenetic regulation is an important mechanism for controlling gene expression and plays key roles in many processes of life, such as development (28,29), as well as in diseases like cancer (30,31). We identified a 102 bp CpG island in the promoter region of the ST8SIA1 gene, and showed that GD3s expression was inversely associated with methylation of the ST8SIA1 synthase gene. Further experiments proved that GD3s expression was increased by demethylation of ST8SIA1. Overexpression of GD3s caused by hypomethylation of ST8SIA1 promoted the development of breast cancer and lowered patients’ survival rates. Taken together, these results showed that GD3s expression was regulated by ST8SIA1 gene methylation and that higher GD3s activity was associated with development of breast cancers.
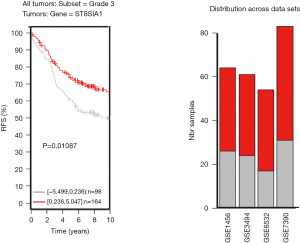
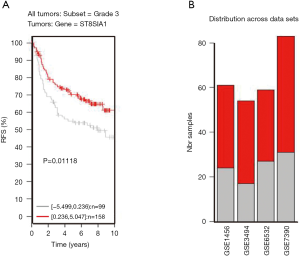
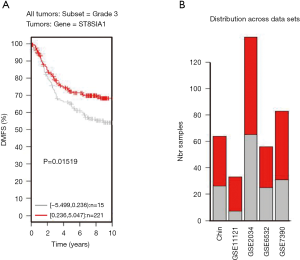
TNBC patients have a poor prognosis and lower OS compared to patients with luminal breast cancer. This may be a result of GD3s over-expression that increases cell proliferation, migration, invasion and colony formation of tumor cells. Higher GD3s expression was first noted in patients with lower rates of OS, relapse-free survival, and distant metastasis-free survival of all breast cancers at grade 3 (Figures S3-S5). GD3s may be used as a biomarker for staging breast cancers and as a potential drug target in the future.
Acknowledgments
Funding: This work was funded by Beijing Natural Science Foundation (7172142) and by National Natural Science Foundation of China (81573454). This work was also funded by Technology Major Projects for “Major New Drugs Innovation and Development” (2018ZX09711001-005-025) and CAMS Innovation Fund for Medical Sciences (2016-I2M-3-007).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The Institutional Review Board at the John Wayne Cancer Institute/Saint John Health Center Providence Health System, Santa Monica, CA, approved the use of human tissues (MORD-RTPCR-0095).
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Bertucci F, Finetti P, Cervera N, et al. How basal are triple-negative breast cancers? Int J Cancer 2008;123:236-40. [Crossref] [PubMed]
- Yanagisawa M, Liour SS, Yu RK. Involvement of gangliosides in proliferation of immortalized neural progenitor cells. J Neurochem 2004;91:804-12. [Crossref] [PubMed]
- Ryu JS, Ko K, Ko K, et al. Roles of gangliosides in the differentiation of mouse pluripotent stem cells to neural stem cells and neural cells Mol Med Rep 2017;16:987-93. (Review). [Crossref] [PubMed]
- Fenderson BA, Eddy EM, Hakomori S. Glycoconjugate expression during embryogenesis and its biological significance. Bioessays 1990;12:173-9. [Crossref] [PubMed]
- Hakomori S. Glycosylation defining cancer malignancy: new wine in an old bottle. Proc Natl Acad Sci U S A 2002;99:10231-3. [Crossref] [PubMed]
- Ko K, Furukawa K, Takahashi T, et al. Fundamental study of small interfering RNAs for ganglioside GD3 synthase gene as a therapeutic target of lung cancers. Oncogene 2006;25:6924-35. [Crossref] [PubMed]
- Zamfir AD, Serb A, Vukeli Z, et al. Assessment of the molecular expression and structure of gangliosides in brain metastasis of lung adenocarcinoma by an advanced approach based on fully automated chip-nanoelectrospray mass spectrometry. J Am Soc Mass Spectrom 2011;22:2145-59. [Crossref] [PubMed]
- Zeng G, Gao L, Suetake K, et al. Variations in gene expression patterns correlated with phenotype of F-11 tumor cells whose expression of GD3-synthase is suppressed. Cancer Lett 2002;178:91-8. [Crossref] [PubMed]
- Carubia JM, Yu RK, Macala LJ, et al. Gangliosides of normal and neoplastic human melanocytes. Biochem Biophys Res Commun 1984;120:500-4. [Crossref] [PubMed]
- Portoukalian J, Zwingelstein G, Dore JF. Lipid composition of human malignant melanoma tumors at various levels of malignant growth. Eur J Biochem 1979;94:19-23. [Crossref] [PubMed]
- Pukel CS, Lloyd KO, Travassos LR, et al. GD3, a prominent ganglioside of human melanoma. Detection and characterisation by mouse monoclonal antibody. J Exp Med 1982;155:1133-47. [Crossref] [PubMed]
- Furukawa K, Arita Y, Satomi N, et al. Tumor necrosis factor enhances GD3 ganglioside expression in cultured human melanocytes. Arch Biochem Biophys 1990;281:70-5. [Crossref] [PubMed]
- Ahmed M, Goldgur Y, Hu J, et al. In silico driven redesign of a clinically relevant antibody for the treatment of GD2 positive tumors. PLoS One 2013;8:e63359. [Crossref] [PubMed]
- Battula VL, Shi Y, Evans KW, et al. Ganglioside GD2 identifies breast cancer stem cells and promotes tumorigenesis. J Clin Invest 2012;122:2066-78. [Crossref] [PubMed]
- Xu J, Fan W, Tu XX, et al. Neural ganglioside GD2(+) cells define a subpopulation of mesenchymal stem cells in adult murine bone marrow. Cell Physiol Biochem 2013;32:889-98. [Crossref] [PubMed]
- Ho MY, Yu AL, Yu J. Glycosphingolipid dynamics in human embryonic stem cell and cancer: their characterization and biomedical implications. Glycoconj J 2017.765-77. [Crossref] [PubMed]
- Vantaku V, Donepudi SR, Ambati CR, et al. Expression of ganglioside GD2, reprogram the lipid metabolism and EMT phenotype in bladder cancer. Oncotarget 2017;8:95620-31. [Crossref] [PubMed]
- Dobrenkov K, Ostrovnaya I, Gu J, et al. Oncotargets GD2 and GD3 are highly expressed in sarcomas of children, adolescents, and young adults. Pediatr Blood Cancer 2016;63:1780-5. [Crossref] [PubMed]
- Fishman PH, Brady RO. Biosynthesis and function of gangliosides. Science 1976;194:906-15. [Crossref] [PubMed]
- Ohkawa Y, Momota H, Kato A, et al. Ganglioside GD3 Enhances Invasiveness of Gliomas by Forming a Complex with Platelet-derived Growth Factor Receptor alpha and Yes Kinase. J Biol Chem 2015;290:16043-58. [Crossref] [PubMed]
- Furukawa K, Kambe M, Miyata M, et al. Ganglioside GD3 induces convergence and synergism of adhesion and hepatocyte growth factor/Met signals in melanomas. Cancer Sci 2014;105:52-63. [Crossref] [PubMed]
- Qu Y, Wang J, Ray PS, et al. Thioredoxin-like 2 regulates human cancer cell growth and metastasis via redox homeostasis and NF-kappaB signaling. J Clin Invest 2011;121:212-25. [Crossref] [PubMed]
- Ray PS, Wang J, Qu Y, et al. FOXC1 is a potential prognostic biomarker with functional significance in basal-like breast cancer. Cancer Res 2010;70:3870-6. [Crossref] [PubMed]
- Sarkar TR, Battula VL, Werden SJ, et al. GD3 synthase regulates epithelial-mesenchymal transition and metastasis in breast cancer. Oncogene 2015;34:2958-67. [Crossref] [PubMed]
- Yeh SC, Wang PY, Lou YW, et al. Glycolipid GD3 and GD3 synthase are key drivers for glioblastoma stem cells and tumorigenicity. Proc Natl Acad Sci U S A 2016;113:5592-7. [Crossref] [PubMed]
- Liang YJ, Wang CY, Wang IA, et al. Interaction of glycosphingolipids GD3 and GD2 with growth factor receptors maintains breast cancer stem cell phenotype. Oncotarget 2017;8:47454-73. [PubMed]
- Soubry A. Epigenetics as a Driver of Developmental Origins of Health and Disease: Did We Forget the Fathers? Bioessays 2018. [Crossref] [PubMed]
- Canovas S, Ross PJ, Kelsey G, et al. DNA Methylation in Embryo Development: Epigenetic Impact of ART (Assisted Reproductive Technologies). Bioessays 2017. [Crossref] [PubMed]
- Pasculli B, Barbano R, Parrella P. Epigenetics of Breast Cancer: biology and clinical implication in the era of Precision Medicine. Semin Cancer Biol 2018;51:22-35. [Crossref] [PubMed]
- Okugawa Y, Grady WM, Goel A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology 2015;149:1204-25.e12. [Crossref] [PubMed]



