
Identification of genes and pathways related to atherosclerosis comorbidity and depressive behavior via RNA-seq and bioinformation analysis in ApoE−/− mice
Introduction
Atherosclerosis (AS) is a chronic pathological condition involving various processes including vascular endothelial dysfunction, inflammatory cell infiltration, abnormal vascular smooth muscle proliferation and migration, lipid peroxide deposition, and vascular matrix changes (1). According to the World Health Organization, diseases with AS as the main pathological feature, such as coronary heart disease (CHD) and ischemic stroke, are high in severity on the disease spectrum (2). Depression is a devastating disease characterized by persistent low mood, lack of pleasure, exhaustion, and has high rates of morbidity, disability, high costs, and even suicide. Moreover, epidemiological studies have confirmed the remarkably high comorbidity between AS and depression (3-5).
Current research suggests that the underlying causes of CHD and depression may be vascular endothelial dysfunction, inflammation, abnormal autonomic nervous system regulation, neuroendocrine-immune network disorder, along with other factors (6). However, inflammation is the main driver of these pathological processes. Patients with CHD and comorbid depression have elevated levels of pro-inflammatory cytokines in the circulating blood, including interleukin (IL)-1β, IL-6, and tumor necrosis factor (TNF)-α (7). Peripheral innate and adaptive immune molecule activation can interact with the central nervous, immune system (8). Toll-like receptors (TLRs) on macrophage-like cells in the choroid plexus may also produce pro-inflammatory cytokines to activate the microglial cells in the central nervous system to respond to the pro-inflammatory cytokines derived from peripheral circulation (9). Despite the above research, the mechanism of coronary atherosclerotic heart disease comorbidity with depression is still poorly understood.
An ApoE−/− mouse is a widely used animal model of AS (10). In this study, the self-state and behavioral changes of ApoE−/− mice were observed and compared with their wild-type mice to determine whether they had depression-like behavioral changes. Furthermore, the RNA-seq method was used to detect the differentially expressed genes (DEGs) in the hippocampus and prefrontal cortex in mice. The function of DEGs was analyzed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations to identify the significant regulatory genes and signal pathways potentially related to the pathogenesis of AS comorbidity with depression. We hope to lay a foundation that can elucidate the pathogenesis of AS comorbidity with depression.
Methods
Animals
Twelve C57BL/6 mice (8±20 g, male, SPF grade) and twelve ApoE−/− mice (8 weeks old, 20±2 g, male, SPF) were purchased from Nanjing Qingzilan Technology Co., Ltd. [animal license number: SCXK (Su) 2016-0010] and the Experimental Animal Center of the Fourth Military Medical University [animal license number: SCXK (Army) 2012-0007], respectively. They were bred in a clean level environment (22±1 °C) with a humidity of 55%±5%, had feed and water provided ad libitum under a daily 12-h light/dark cycle. After one 1 week of adaptive diet feeding, they were weighed and divided into cages according to the statistical random grouping principle. The groups were defined as follows: control group, C57 mouse + ordinary diet, and AS model group, ApoE−/− mouse + high-fat diet. All animals were kept in strict accordance with the National Laboratory Animal Management Regulations and guidelines of the Animal Feeding and Ethics Committees of the Experimental Animal Center of Nanjing University of Chinese Medicine (Laboratory Animal Ethics No.: ACU171104).
Experimental design
Bodyweight and coat state score
The mice were weighed every 2 weeks, and the coat state was assessed every 2 weeks as a marker of the progression of the depressive syndrome. Specifically, the scoring sites included the head, neck, back, abdomen, and back claw (n=5). The possible scores were 0 or 1. A 0 score meant the hair of the mouse was clean, neat, and smooth, whereas a 1 score meant it was messy, fluffy, and greasy. Two experimenters assigned the total score of each mouse according to the scoring criteria. The grader did not know the grouping and drug administration of the mice (11).
Sucrose preference test
A bottle of 1% sucrose water and a bottle of pure water were made freely available to the mice for 24 h. The position of the two bottles was changed every 12 h, and the food intake was not restricted when adapting. Then, the amounts of water and sucrose solution consumed in each group were weighed. The formula for calculating the sugar preference was as follows: sugar preference ratio (%) = sucrose consumption/(sucrose consumption + pure water consumption) ×100% (12,13).
Open-field test
This test was performed in a manner similar to that described previously (14) and repeated 3 times (1st, 7th, and 15th weeks) to test the changes in the activity pattern with increasing experimental time (15). To study spontaneous locomotion, the mice were placed in an open rectangular plastic apparatus (25×25×35 cm3) with 4 squares. The spontaneous activities of the mice were observed and recorded for 5 min using the TopScan system. Then, the horizontal spontaneous motion distances (total movement distance, central area distance, central area time/total time) of the mice were systematically analyzed using a small animal behavior trajectory analysis system.
Light/dark test
The light/darkfield device consisted of 2 boxes of plexiglass (20×20×14 cm3). One of the boxes was darkened. The lamp from the desk lamp, about 10 cm above the other box, provided illumination for the room. An opaque plastic tunnel (5×7×10 cm3) separated the black box from the lighting box. At the beginning of the experiment, the mouse was placed in the illuminated box, facing the tunnel. The device was equipped with an infrared beam and sensor to measure the relevant parameters for 5 min (12).
Tail suspension test
The mice were hung upside down in a white plastic box (25×25×35 cm3). The nose tip of each mouse was held 50 cm above the floor. The mice were suspended from their tails 1 cm from the end using a medical adhesive tape. Each mouse was monitored for the duration of 6 min, which comprised an initial 2 min and a subsequent 4 min of acclimatization and testing periods, respectively (16). During the test, the mice were completely separated from each other to avoid possible visual and auditory interference
Enzyme-linked immunosorbent assays in the levels of the pro-inflammatory factors and plasma lipids
At the end of the behavioral experiment, fasting was performed for 8 h, and 5% sucrose water was given to supply energy. The blood was taken from the posterior venous vein and placed in a 1.5-mL Eppendorf (EP) tube. After standing in a refrigerator at 4 °C for 1 h, the plasma samples were obtained after centrifugation at 3,000 rpm at 4 °C for 15 min. The levels of the pro-inflammatory factors (including IL-1β, IL-6, and TNF-α) and lipids [total cholesterol (TC), triglycerides (TG), and LDL-cholesterol (LDL-C)] were detected using the enzyme-linked immunosorbent assay kits mentioned earlier according to the manufacturers’ instructions.
Oil Red O and hematoxylin and eosin (HE) staining
After anesthesia with pentobarbital sodium, the mice were fixed on the dissection table, and the chest cavity was fully exposed. They were perfused through the left ventricle with cold PBS (pH, 7.2), and then, after being infused with cold 4% paraformaldehyde, the base of the ascending aorta was carefully separated to the brachial artery. Subsequently, the entire aortas of each group were stained with oil red O for the lipid accumulation analysis. The lesion areas were analyzed by Image-J Software.
HE staining of the pathological sections of the aortic root tissue was performed according to the manufacturer’s instructions. Furthermore, the morphological changes of the endothelium, smooth muscle cells, and foam cells on the aortic arch tissues were observed by light microscopy.
Total RNA extraction
Using the method of cervical dislocation, all the mice were sacrificed; the hippocampus and prefrontal cortex tissues obtained from the sacrificed mice were rapidly separated using ice. Further, the tissues were frozen using liquid nitrogen and stored at a temperature of −84 °C.
From the frozen tissues, total RNA was extracted using Trizol (Invitrogen, Carlsbad, CA, USA). The extraction was performed according to the instruction manual. Next, using a 2-mL tube and liquid nitrogen, nearly 30 mg of the tissues were ground into powder. The obtained powder was then homogenized for 2 min and rested horizontally for 5 min thereafter. The contents were subjected to centrifugation at 12,000 ×g and 4 °C for 5 min; also, the supernatant was taken into an EP tube and mixed with 0.3 mL chloroform/isoamyl alcohol (24:1); the contents were vigorously shaken for 15 s, followed by centrifugation at 12,000 ×g and 4 °C for 10 min; after this step, the upper aqueous phase containing RNA was transferred into a new tube, and an equal volume of supernatant of isopropyl alcohol was mixed with the aqueous phase. The contents were then centrifuged at 13,600 rpm and 4 °C for 20 min. The supernatant obtained after the centrifugation was discarded, and the RNA pellet was washed with 1-mL 75% ethanol twice; the resulting contents were then centrifuged at 13,600 rpm and 4 °C for 3 min, and the residual ethanol was separated. The remaining pellet was air-dried in a biosafety cabinet for 5–10 min. To dissolve the RNA, nearly 25–100 µL of diethylpyrocarbonate (DEPC)-treated water was added to the dried pellet. The resulting total RNA was qualified and quantified using a NanoDrop and Agilent 2100 bioanalyzer (Thermo Fisher Scientific, MA, USA).
mRNA library construction
Double- and single-stranded DNA in the total RNA was digested using DNase I. This led to the production of magnetic beads that were further purified for recovering the reaction products. To remove the RNA, RNase H or Ribo-Zero method (human, mouse, plants) (Illumina, USA) was employed. Consequently, purified mRNA was obtained and eventually fragmented into small pieces using a fragment buffer at a suitable temperature. Later, First-Strand Reaction System, along with polymerase chain reaction (PCR), was used for producing first- and second-strand cDNA. The magnetic beads were then utilized for purification of the reaction product. A-Tailing Mix and RNA Index Adapters were added by incubation to carry out end repair. Further, on PCR, the cDNA fragments with adapters were amplified, and the products obtained were then purified using Ampure XP Beads. For quality control purposes, the cDNA Library was validated on the Agilent Technologies 2100 bioanalyzer. The aforementioned double-stranded PCR products were subjected to heat denaturation, followed by circularization using the splint oligo sequence. The final cDNA library was constructed by formatting the single-strand circular DNA (ssCir DNA) and amplified with phi29 (Thermo Fisher Scientific, MA, USA) for generating DNA nanoballs (DNBs) that comprised more than 300 copies of one molecule. These DNBs were transferred into the patterned nanoarray. Finally, using the BGISEQ500 platform (BGI-Shenzhen, China), single-end 50 bases reads were successfully generated.
Differential expression gene analysis
The RNA-seq method is based on the Poisson distribution. In this study, the gene expression levels were evaluated using R software package-based MA-plot, which is a statistical analysis tool used widely for the detection and visualization of the intensity-dependent ratio of microarray data. M is defined as (log2C1 − log2C2), whereas A is defined as (log2C1 + log2C2)/2 (17). For the aforementioned methods, P values were calculated and adjusted for each gene. Genes with a false discovery rate threshold of >2 and a Q-value of ≤0.001 were screened as significant DEGs (18,19).
Functional annotation of DEGs
A classification and enrichment analysis of the GO function was performed for DEGs on the basis of the results of the DEGs. GO was divided into three major functional categories: molecular function, cellular component, and biological process. We further classified and enriched the three functional categories of DEGs separately.
The KEGG is a knowledge base for systematic analysis of gene function, linking genomic information and functional information (http://www.genome.jp/kegg/). The pathway is based on the relevant knowledge to determine the connections of the components of the path in a specific language format. Based on the results of DEGs, we performed KEGG biological pathway classification and enrichment analysis.
Statistical analysis
The results were expressed as mean ± standard error of means (SEM). The analysis was completed using SPSS 22 software (IBM Corp., USA). In addition, the comparisons between the control and model groups were analyzed using Student’s t-test, whereas the body weight, coat score, sugar preference, and the open-field tests were analyzed by the two-way analysis of variance (ANOVA) tests, followed by Tukey’s post hoc test when significant differences were detected. P values <0.05 were considered statistically significant.
Results
ApoE−/− mice fed with High-fat diet gradually developed depression-like behavioral changes
We evaluated the depression-like behavior in high-fat-fed ApoE−/− mice using the body weight, coat color score, sugar preference, tail suspension, and open-field tests. As shown in Figure 1A, the body weights of the model and control groups increased with the prolonged feeding time, but the bodyweight of the model group increased more rapidly. From the 11th week onward, the bodyweight of the model group was significantly different from that of the control group.
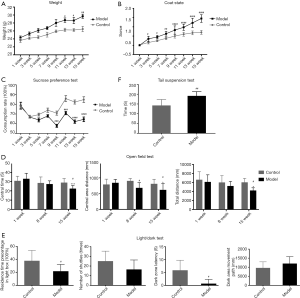
The coat state of the mice was observed continuously every 2 weeks, as shown in Figure 1B. The results showed that the coat color scores of the model and control groups increased with the feeding time, and the coat color score increased faster in the model group, starting from the 3rd week. Moreover, a significant difference in the coat color score between the model and control groups was noted. The back-hair condition of the model group was significantly worse than that of the control group, the combing behavior was significantly reduced, and the coat color score was higher than that of the control group.
The sugar preference test by the ratio of sugar water consumption was used to evaluate whether the euphoria of the experimental mice was missing. As shown in Figure 1C, the saccharide consumption ratio of the model group was lower than that of the control group from the 9th week, and a significant difference was noted.
The open-field experiment judged the state of depression by evaluating the spontaneous movement of the mice. As shown in Figure 1D, during the experiment, the 1st, 7th, and 15th weeks were selected to compare the central zone distance of the two groups of mice, central zone residence time, and spontaneous movement index of the total distance. Consequently, no significant difference was noted in the spontaneous exercise index between the two groups in the first week. In the 7th week, no difference in the distance was identified between the model and control groups except the central area (P<0.05). Also, no significant difference was noted between the other total distances and the central time. At the end of the 15th week of the experiment, compared with the blank, the central time, central distance, and total distance of the model group movement were reduced relative to the control, and the difference was statistically significant (P<0.05).
In the light-dark test (Figure 1E), we found that mice in the model group spent less time in the lightbox than those in the control group, showing significant differences (P<0.01). As expected, mice in the model group displayed less latency behavior in the dark box than those in the control group (P<0.05).
The tail suspension experiment responded to depression by detecting the inactivity time of the inverted mouse. As shown in Figure 1F, experiments were performed at the 15th week of the experimental endpoint. The immobility time of the model group was significantly higher than that of the control group, and the difference was statistically significant.
Based on the above experimental results, it can be seen that with the prolongation of high-fat feeding time, ApoE−/− mice gradually showed depression-like behavior, while the control group mice showed no depression-like behavioral changes.
Blood lipids, inflammatory factors, and atherosclerotic plaque formation in ApoE−/− mice fed with a high-fat diet for 15 weeks
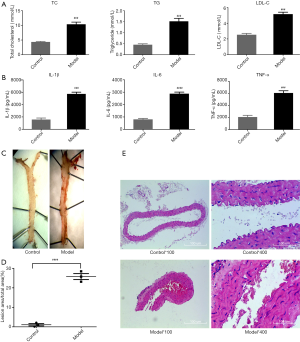
As shown in Figure 2A, the levels of the blood lipids TC, TG, and LDL-C were significantly increased in the model group compared with the control group, and the inflammatory factors IL-1β, IL-6, and TNF-α in the serum were also significantly increased (Figure 2B). The mouse thoracic aorta oil red O staining (Figure 2C,D) and HE staining (×100, 100-fold; ×400, 400-fold, Figure 2E) indicate that ApoE−/− mice fed with a high-fat diet had significant atherosclerotic plaque formation after 15 weeks.
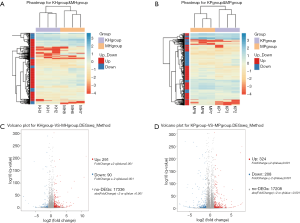
An overview of the RNA-SEQ data
A total of 12 samples were tested using the BGISEQ-500 platform, 6 from the control group, and 6 from the model group, with an average yield of 23.97 M data per sample. The average alignment rate of each sample and ratio of the pair of genes was 95.31% and 69.96%, respectively. The raw data of the sequencing was filtered. Based on the gene expression levels of each sample, we detected DEGs between the mouse hippocampus and the prefrontal cortex (sample group). This project used the RNA-seq method for differential detection, expressing the calorimetry for each group of DEGs, displaying the distribution of DEGs using a volcano plot, and performing GO and pathway function analyses on DEGs.
Analyses of DEGs in the hippocampus and prefrontal cortex
To find the DEGs in the hippocampus of the control and model groups, DEG screening was conducted, and a total of 17,717 genes were detected. Moreover, 291 genes were upregulated, and 90 genes were downregulated. Similarly, to find DEGs in the prefrontal cortex of the control and model groups, a total of 17,208 genes were detected. Among them, 324 genes in the model were compared with the control group, and 208 genes were downregulated. The expression calorimetry maps of DEGs in the hippocampus and prefrontal cortex can be seen in Figure 3A,B. A volcano plot was used to display the distribution of DEGs in the hippocampus and prefrontal cortex, as shown in Figure 3C,D respectively.
GO function analysis of DEGS in the hippocampus and prefrontal cortex
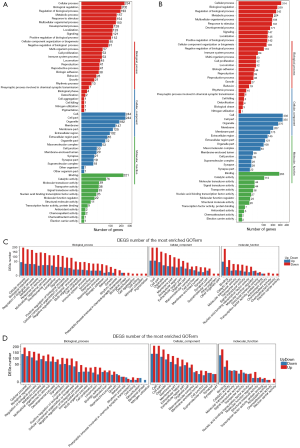
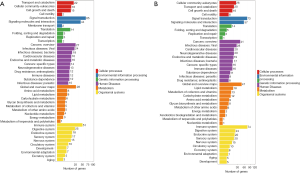
To characterize the functional distribution of genes on a macro-level, the GO classification and functional enrichment analysis were performed to determine the DEG function in the hippocampal and prefrontal cortex samples of the model and control groups. The GO function classification results of the hippocampus and prefrontal cortex tissues are shown in Figure 4A,B, and the upregulations of DEGs corresponding to GO function are shown in Figure 4C,D. According to the results of GO annotation, the results of DEG detected in the hippocampus, and prefrontal cortex was similar in terms of molecular function, cellular components, and biological processes.
For biological process, the “cell process” (234 DEGs for the hippocampus and 314 DEGs for the prefrontal cortex) was the most highly represented GO term, followed by “biological regulation” (202 DEGs for the hippocampus and 259 DEGs for the forehead cortex), and the “behavior” team had 28 DEGs for the hippocampus and 19 DEGs for the prefrontal cortex.
For the cellular component, the highest numbers of DEGs were “cell” and “cell part” for the hippocampus (244 DEGs); however, the highest number of DEGs for the prefrontal cortex was “cell” (336 DEGs), followed by “cell part” term (335 DEGs).
For molecular function, “binding” (221 DEGs for the for 285 DEGs for the hippocampus and prefrontal cortex) was the most highly represented the GO term, followed by “catalytic activity” (76 DEGs for the hippocampus and 113 DEGs and for the prefrontal cortex).
As shown in Figure 4C, the number of upregulated genes in the hippocampus was higher than that in the downregulated genes. As shown in Figure 4D, the DEGs in the prefrontal cortical tissue had more reduction of GO term than the upper one. For instance, in DEGs of behavior in the biological process, the up-regulated genes were higher in the hippocampus than in the down-regulated genes, while the down-regulated genes were higher in the prefrontal cortex than in the up-regulated genes.
KEGG functional annotation of DEGs in the hippocampus and prefrontal cortex
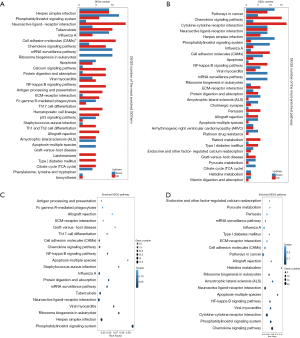
According to the results of differential gene detection in the hippocampus and prefrontal cortex, we performed the KEGG biological pathway classification and enrichment analysis for the DEGs. The results of pathway classification are shown in Figure 5A,B. The results of pathway enrichment are also shown in Figure 6A,B, and the statistics of pathway enrichment of DEGs are shown in Figure 6C,D.
Whether it was for the hippocampus or for the prefrontal cortex, the immune system and signal transduction pathways had the greatest representation of DEGs. The immune system is an organismal system, and signal transduction is part of environmental information processing.
Only 1 DEG was mapped to the metabolism of the terpenoids and polyketides pathway in the hippocampus. For the prefrontal cortex, there was only 1 DEG mapped to transcription, metabolism of terpenoids, and nucleotide metabolism.
As shown in Figure 6C,D, the largest value of a richer factor for the hippocampus was the phosphatidylinositol signaling system, while for the prefrontal cortex, the largest value of a richer factor was for histidine metabolism.
The smallest value of Q value for the hippocampus was also the phosphatidylinositol signaling system, while for the prefrontal cortex, the smallest value of Q value was for the chemokine signaling pathway.
As shown in Figure 6A,C, for the DEGs of the hippocampus, the pathway with the most upregulated gene expressions is neuroactive ligand-receptor interaction. The NF-kappa B signaling, Th17 cell differentiation, and Th1 and Th2 cell differentiation pathways only had upregulated gene expression with no downregulated expression of genes.
For the DEGs of the prefrontal cortical DEGs, the pathway with the most upregulated gene expression was the cytokine–cytokine receptor interaction pathway. The DEGs associated with the NF-kappa B signaling pathway was mainly upregulated.
Discussion
The relationship between AS and depression is fairly well acknowledged. Still, the current understanding of the mechanism linking AS and depressive disorder is in need of more extensive clarification. While research relevant to this issue has been undertaken, the majority of these studies are based on clinical observations, and no experimental animal model exploring the link between AS and depression has been explored. In this experiment, ApoE−/− mice, which are gene-deficient mice used for classic AS models, were fed with a high-fat diet to induce the pathological changes of AS, and subsequently evaluated for depressive behavior. The findings of this experiment revealed that AS model mice had depression-like behavioral changes.
The changes were characterized by a gradual increase in body weight and a decrease in skin gloss and cleanliness in AS-compressed mice. In the open-field experiment, the horizontal movement distance and central area residence time were reduced. Furthermore, the sucrose consumption rate of the model group was higher than that of the control group, and it was also lower than the fluctuation rate of the sucrose consumption rate of the control group as the experimental time increased. Furthermore, in the sucrose preference test, the sucrose consumption rate in both the model group and the control group showed fluctuation. Compared with the control group, the sucrose consumption rate of the model group decreased. In the light/dark box experiment, the number of shuttles in model group was reduced, the dwell time in the dark field of the model group was longer, and the moving distance was lower than that in the control group. The tail suspension experiment was performed at the end of the study. Consequently, the model group had significantly more immobility time than the control group.
Compared with the blank group, the levels of TG, TC, and LDL-C in the circulating blood of the mice in the model group were significantly increased, and the classical inflammatory factors IL-1β, IL-6, and TNF-α were also higher than those in the blank group.
The gross aortic red O staining and plaque area statistics of the aorta showed a significant pathological change in AS in the aorta of the model group. HE staining results also supported the conclusion of atherosclerotic lesions being present in the arteries of mice, and that eosinophil infiltration occurred in plaque lesions.
Based on the aforementioned results, the present experiment demonstrated that ApoE-/- mice fed a high-fat diet for 15 weeks, showed higher blood lipids, elevated chronic inflammatory factors in circulating blood, and obvious pathological changes in aortic AS compared with those in the control group (C57 mice).
AS is a common chronic pathological condition in mammals (20), and long-term physical illness can easily lead to depression and mood disorders (21). To clarify the pathogenesis of AS comorbidity with depressive behavior in ApoE−/− mice, RNA-seq and bioinformatics were used for the analysis and annotation of DEGs.
For the DEGs from the hippocampus area, the GO annotation analysis showed that the cellular components of the DEGs mainly focused on various membrane structures. (including the plasma membrane part, external side of the plasma membrane, an intrinsic component of the plasma membrane, cell periphery, plasma membrane, cell surface, an integral component of the plasma membrane, side of the membrane, plasma membrane region, basolateral plasma membrane).
The GO annotation of DEGs from the hippocampus tissue showed that G-protein-coupled receptor binding, CCR7 chemokine receptor binding, receptor binding, CCR chemokine receptor binding, chemokine activity, neuropeptide hormone activity, calcium ion binding, chemokine receptor binding, transporter activity, and neuropeptide receptor binding were the main molecular functions.
The biological processes of DEGs were mainly related to the neuroimmune system (response to external stimulus, positive regulation of ERK1 and ERK2 cascade, neuropeptide signaling pathway, positive regulation of synaptic transmission, cholinergic), especially the immune-inflammatory signaling pathway (the regulation of neutrophil chemotaxis, the monocyte chemotaxis, lymphocyte chemotaxis. and myeloid dendritic cell chemotaxis). This difference in biological processes was also reflected in the effects on feeding behavior and regulation of saliva secretion.
The KEGG annotation results demonstrated that the signaling pathway involved in DEGs was the phosphatidylinositol signaling system, which primarily regulated the environmental information processing and signal transduction. Calmodulin-dependent kinase II, a protein of the brain neuron synapses in the mammalian hippocampus, is abundant and involved in memory formation. The results of the present study indicate that gene mutations in ApoE−/− mice that regulate this protein had a significant decrease in memory function. In addition, genes that regulate neuronal ligand-receptor interactions and signaling pathways for protein digestion and absorption were the results of KEGG-annotated DEGs.
The results of functional annotation of DEGs revealed that from the prefrontal cortex area, the downregulated DEGs were mainly enriched in the biological process terms associated with behavior, localization, locomotion, and reproductive process and pathways associated with the regulation of neuron projection development, homophilic cell adhesion via plasma membrane adhesion molecule, calcium ion-regulated exocytosis of neurotransmitter, cerebral cortex GABAergic interneuron fate commitment, central nervous system vasculogenesis, chemoattraction of dopaminergic neuron axon, neuron projection morphogenesis, and neuron development and metabolism.
The upregulated DEGs were mainly enriched in the BP terms associated with immune system process, the presynaptic process involved in chemical synaptic transmission, rhythmic process, and pathways associated with TLR-4 binding, NF-kappa B signaling pathway, cellular response to TNF-α, cytokine-cytokine receptor interaction, CCR7 chemokine receptor binding, and cytokine activity.
Most of the evidence from preclinical and clinical studies suggest that psychiatric illnesses, particularly depression or major depressive disorder (MDD), are associated with inflammatory processes (22). Some scholars believe that depression is a microglia disease of the central nervous system (23). When the activation of the peripheral immune system continues unabated, as in cancer or AS (24,25), the ensuing immune signaling to the brain can lead to an exacerbation of sickness and the development of symptoms of depression in vulnerable individuals. The brain monitors the peripheral innate immune responses (26), while locally produced cytokines activate primary afferent nerves, such as the vagus nerve during AS and visceral infection, and transmit inflammatory immune signals from the periphery to the brain (27). In the present research, most of the upregulated DEGs from both the hippocampus and prefrontal cortex were of the cytokine-cytokine receptor interaction, chemokine receptor binding and cytokine activity, and NF-kappa B signaling pathway. The high expression of these inflammatory pathway genes may lead to the inhibition of synaptic signal transduction (28), inhibition of hippocampal neuronal regeneration (29), decreased secretion of neurotrophic factors (30), and inhibition of dopaminergic signaling pathway (31).
In summary, in ApoE−/− mice fed with a high-fat diet, in addition to the basic signs of AS, the RNA-seq and bioinformatics analysis revealed the high expression of inflammatory chemokine genes in the hippocampus and prefrontal cortex areas. These areas also showed a reduction in the regulation of movement, feeding, and reproduction of gene expression, which could reasonably explain the gene regulation mechanism of depression-like behavioral changes in ApoE−/− mice. The results of this experiment support the notion that ApoE−/− mice can be used as an animal model of AS with depression-like changes.
Although bioinformatics technologies have the potential to identify and validate candidate agents for critical diseases, certain limitations remain in this study. Firstly, the sample size for microarray analysis was small, which might have caused a high rate of false-positive results. Secondly, this study lacked real-time RNA or Western blot experiment verification. Further genetic and experimental studies with a larger sample size are still required in the future to confirm the results.
Despite these limitations, the data of the present study provide a comprehensive bioinformatics analysis of the DEGs and pathways that may be involved in AS comorbidity with depressive behavior in ApoE−/− mice fed with a high-fat diet. The findings of the present study may contribute to the development of a new model mouse useful for further understanding the underlying molecular mechanisms of AS comorbidity with depression.
Conclusions
These results indicate that when ApoE−/− mice were fed with high-fat diets continuously for 15 weeks, depression-like behavioral changes occurred in the mouse model of atherosclerotic pathology. The RNA-seq, combined with bioinformatics analysis, showed that this AS comorbidity with depressive behavior was associated with the high expression of inflammation-related genes and pathways in the hippocampus and prefrontal cortex.
Acknowledgments
We thank Linran Han and Jianjun Xu for their help in animal experiments.
Funding: This work was supported by the National Natural Science Foundation of China (Nos. 81173591 and 81373855). It was also supported by funding from the Priority Academic Program Development of Jiangsu Higher Education Institutions (integration of Chinese and western medicine) and by the high-level talents of Nanjing University of Chinese Medicine.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All of the protocols were approved by the Ethics Committee of the Laboratory Animal Center of the Nanjing University of Chinese Medicine (Laboratory Animal Ethics No. ACU171104).
References
- Weber C, Noels H. Atherosclerosis: current pathogenesis and therapeutic options. Nat Med 2011;17:1410-22. [Crossref] [PubMed]
- WHO. The top 10 causes of death. 2018. Available online: http://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death
- Halaris A. Inflammation-Associated Co-morbidity Between Depression and Cardiovascular Disease. Curr Top Behav Neurosci 2017;31:45-70. [Crossref] [PubMed]
- Konrad M, Jacob L, Rapp MA, et al. Depression risk in patients with coronary heart disease in Germany. World J Cardiol 2016;8:547-52. [Crossref] [PubMed]
- Seldenrijk A, van Hout HP, van Marwijk HW, et al. Carotid atherosclerosis in depression and anxiety: associations for age of depression onset. World J Biol Psychiatry 2011;12:549-58. [Crossref] [PubMed]
- Carney RM, Freedland KE. Depression and coronary heart disease. Nat Rev Cardiol 2017;14:145-55. [Crossref] [PubMed]
- Golimbet VE, Volel BA, Korovaitseva GI, et al. Association of inflammatory genes with neuroticism, anxiety and depression in male patients with coronary heart disease. Zh Nevrol Psikhiatr Im S S Korsakova 2017;117:74-9. [Crossref] [PubMed]
- Haapakoski R, Ebmeier KP, Alenius H, et al. Innate and adaptive immunity in the development of depression: An update on current knowledge and technological advances. Prog Neuropsychopharmacol Biol Psychiatry 2016;66:63-72. [Crossref] [PubMed]
- Boche D, Perry VH, Nicoll JA. Review: activation patterns of microglia and their identification in the human brain. Neuropathol Appl Neurobiol 2013;39:3-18. [Crossref] [PubMed]
- Palinski W, Ord VA, Plump AS, et al. ApoE-deficient mice are a model of lipoprotein oxidation in atherogenesis. Demonstration of oxidation-specific epitopes in lesions and high titers of autoantibodies to malondialdehyde-lysine in serum. Arterioscler Thromb 1994;14:605-16. [Crossref] [PubMed]
- Gosselin T, Le Guisquet AM, Brizard B, et al. Fluoxetine induces paradoxical effects in C57BL6/J mice: comparison with BALB/c mice. Behav Pharmacol 2017;28:466-76. [Crossref] [PubMed]
- Culig L, Surget A, Bourdey M, et al. Increasing adult hippocampal neurogenesis in mice after exposure to unpredictable chronic mild stress may counteract some of the effects of stress. Neuropharmacology 2017;126:179-89. [Crossref] [PubMed]
- Kurhe Y, Mahesh R. Pioglitazone, a PPARγ agonist rescues depression associated with obesity using chronic unpredictable mild stress model in experimental mice. Neurobiol Stress 2016;3:114-21. [Crossref] [PubMed]
- Mutlu O, Gumuslu E, Ulak G, et al. Effects of fluoxetine, tianeptine and olanzapine on unpredictable chronic mild stress-induced depression-like behavior in mice. Life Sci 2012;91:1252-62. [Crossref] [PubMed]
- Shaltiel G, Maeng S, Malkesman O, et al. Evidence for the involvement of the kainate receptor subunit GluR6 (GRIK2) in mediating behavioral displays related to behavioral symptoms of mania. Mol Psychiatry 2008;13:858-72. [Crossref] [PubMed]
- Zhou Y, Wang X, Zhao Y, et al. Elevated Thyroid Peroxidase Antibody Increases Risk of Post-partum Depression by Decreasing Prefrontal Cortex BDNF and 5-HT Levels in Mice. Front Cell Neurosci 2017;10:307. [Crossref] [PubMed]
- Wang L, Feng Z, Wang X, et al. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010;26:136-8. [Crossref] [PubMed]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - A Practical And Powerful Approach To Multiple Testing. J R Stat Soc Series B Stat Methodol 1995;57:289-300.
- Storey JD, Robert T. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 2003;100:9440-5. [Crossref] [PubMed]
- Tall AR, Yvan-Charvet L. Cholesterol, inflammation and innate immunity. Nat Rev Immunol 2015;15:104-16. [Crossref] [PubMed]
- Licinio J, Yildiz B, Wong ML. Depression and cardiovascular disease: co-occurrence or shared genetic substrates? Mol Psychiatry 2002;7:1031-2. [Crossref] [PubMed]
- Miller AH, Raison CL. The role of inflammation in depression: from evolutionary imperative to modern treatment target. Nat Rev Immunol 2016;16:22-34. [Crossref] [PubMed]
- Yirmiya R, Rimmerman N, Reshef R. Depression as a Microglial Disease. Trends Neurosci 2015;38:637-58. [Crossref] [PubMed]
- Ciobanu LG, Sachdev PS, Trollor JN, et al. Differential gene expression in brain and peripheral tissues in depression across the life span: A review of replicated findings. Neurosci Biobehav Rev 2016;71:281-93. [Crossref] [PubMed]
- Raison CL, Miller AH. Depression in cancer: new developments regarding diagnosis and treatment. Biol Psychiatry 2003;54:283-94. [Crossref] [PubMed]
- Galea I, Bechmann I, Perry VH. What is immune privilege (not)? Trends Immunol 2007;28:12-8. [Crossref] [PubMed]
- Buller KM. Role of circumventricular organs in pro-inflammatory cytokine-induced activation of the hypothalamic-pituitary-adrenal axis. Clin Exp Pharmacol Physiol 2001;28:581-9. [Crossref] [PubMed]
- Wohleb ES, Franklin T, Iwata M, et al. Integrating neuroimmune systems in the neurobiology of depression. Nat Rev Neurosci 2016;17:497-511. [Crossref] [PubMed]
- Difato F, Tsushima H, Pesce M, et al. Photonic Therapeutics and Diagnostics VIII - Axonal regeneration of cultured mouse hippocampal neurons studied by an optical nano-surgery system. SPIE Proceedings, SPIE BiOS - San Francisco, California, USA (Saturday 21 January 2012), 2012;8207:102.
- Zhang JC, Yao W, Hashimoto K, et al. Brain-derived Neurotrophic Factor (BDNF)-TrkB Signaling in Inflammation-related Depression and Potential Therapeutic Targets. Curr Neuropharmacol 2016;14:721-31. [Crossref] [PubMed]
- Hopp SC, Royer SE, D’Angelo HM, et al. Differential Neuroprotective and Anti-Inflammatory Effects of L-Type Voltage Dependent Calcium Channel and Ryanodine Receptor Antagonists in the Substantia Nigra and Locus Coeruleus. J Neuroimmune Pharmacol 2015;10:35-44. [Crossref] [PubMed]


