
The genetic contribution of HLA-E*01:03 and HLA-E*01:03-G*01:01 to Posner-Schlossman syndrome in southern Chinese
Introduction
Posner-Schlossman syndrome (PSS), also known as a glaucomatocyclitic crisis, was first reported by Posner and Schlossman in 1948 (1). PSS is a secondary glaucoma, which occurs in young adults. PSS is a self-limiting and recurrent eye disorder, and 24.3% of patients have more than 2 episodes per year, up to 12 episodes in some cases (2). The main clinical manifestations of PSS are the elevated intraocular pressure (IOP) in one eye with keratic precipitates (KPs). Normally, symptoms in most patients can be relieved by using IOP-reducing and anti-inflammatory drugs, but many studies have shown that PSS is not a completely benign disease (3). Long-term recurrent PSS can lead to optic nerve damage and a reduced number of corneal endothelial cells. Permanent visual impairment can develop in severe cases. Anti-glaucoma surgery is needed in some patients with PSS because of inadequate disease control (3,4).
The human leukocyte antigen (HLA) gene is located in the short arm of chromosome 6 (6p21.31), with a total length of about 3.6 Mb. It is rich in polymorphism and takes part in immune response and the regulation of immune function (5). HLA gene polymorphisms are associated with a variety of eye diseases, such as uveitis, glaucoma, Graves’ ophthalmopathy, and the transplant rejection after corneal transplantation (6-10). The polymorphisms of the classical HLA-Ia and HLA-II genes have been associated with PSS (11-13). The study of Hirose et al. [1985] first reported that the HLA-Bw54 and HLA-Bw54-Cw1 haplotype had a significantly higher frequency in Japanese patients with PSS than in normal controls (11). In two recent studies of the southern Chinese population, Zhao et al. found that HLA-C*14:02, HLA-A*11:01-C*14:02 and HLA-B*51:01-C*14:02 were associated with increased risk of PSS, while HLA-B*13:01, HLA-DPA1*02:01, HLA-DPB1*14:01, HLA-DPB1*17:01, HLA-B*13:01-C*03:04 and HLA-DPB1*14:01-DPA1*02:01 might be protective factors for PSS (12,13).
Non-classical HLA-Ib (HLA-E and HLA-G) molecules play an essential inhibitory role in innate and adaptive immunity (14,15). The receptors can recognize HLA-E molecules on natural killer (NK) cells and some cytotoxic lymphocytes (CTL) (CD94/NKG2 receptor) to regulate cytotoxic activity (15). HLA-G molecules can directly inhibit the function of immune cells (NK cells, CTLs, B cells, and dendritic cells) (14,15). HLA-E and HLA-G gene polymorphisms are closely related to viral infection, rejection after organ transplantation, tumor surveillance, and autoimmune system abnormalities (16-25). However, the correlation between HLA-E and HLA-G gene polymorphisms and the pathogenesis of PSS is still unclear. In this study, we evaluated the association between HLA-E and HLA-G gene polymorphisms and PSS in a southern Chinese Han population.
Methods
Patients and controls
Between December 2015 and December 2018, a total of 97 unrelated PSS patients were recruited from patients attending the Shenzhen Eye Hospital Clinic. The diagnosis of PSS was based on the following criteria (1-4,12,13): (I) single-eye onset in young adults, mild discomfort in the eye, and no significant decrease or slight decrease in visual acuity; (II) elevated IOP with recurrent episodes and mutton-fat KPs; (III) open iridocorneal angle under high IOP without peripheral anterior synechia; (IV) no visual field loss and optic nerve damage in patients with shorter course of disease; and (V) no history of other eye diseases except for refractive error. Ninety unrelated subjects were recruited at the Shenzhen Blood Center from healthy volunteer blood donors with normal IOP and optic discs. Patients and controls were all southern Han Chinese and matched on age, sex, and ethnicity. The study protocol was approved by the Ethics Committee of Shenzhen Eye Hospital and was in accordance with the tenets of the Declaration of Helsinki and its later amendments or comparable ethical standards. Written informed consent was obtained from all study participants.
DNA extraction
DNA was extracted from peripheral blood samples of all participants using the MagCore nucleic acid extraction kit according to the instructions of the manufacturer (Promega Corporation, Madison, WI, USA).
Polymerase chain reaction (PCR) amplification
The full-length sequences of HLA-E and -G were amplified by long-range high-fidelity PCR. PCR amplification was performed in a 20 µL reaction system, including 0.5 µL pfuUltraTM Fusion HS DNA polymerase, 2 µL of 10× pfuUltraTM Rxn Buffer, 1 µL of dNTP (2.5 mmol/L) mixture, 0.5 µL of each PCR primer (10 µmol/L, Table 1), 14.5 µL ddH2O, and 1 µL genomic DNA.

Full table
The PCR products were purified using a magnetic bead reagent before the sequencing reaction to remove non-specific products generated during PCR amplification. Eighteen µL Mag-Bind EZ Pure magnetic beads were added to each well and mixed thoroughly. After standing for 10 min, the plate was placed on a magnetic stand. After adsorbing on the magnetic beads for 10 min, the supernatant was discarded. 200 µL of 70% ethanol was used to wash twice for 1 min. After standing at room temperature for 20 min for the ethanol completely evaporated, 30 µL of ddH2O was added to each well to collect the purified products.
Selection of polymorphisms and genotyping
Because the HLA-E*01:01 allele and the HLA-E*01:03 allele are different at the rs1264457 site of the third exon, exon 3 of the HLA-E gene was selected for sequencing (26,27). Since there are 43 single nucleotide polymorphism sites in the HLA-G gene according to the IMGT/HLA database (http://www.ebi.ac.uk/ipd/imgt/hla/, Release 3.36.0, 2019 April), the coding region of the HLA-G gene was sequenced. The sequencing primers were listed in Table 2.

Full table
The sequencing reaction products (10 µL) were purified by adding 2.5 µL NaOAc/EDTA and 50 µL of 80% ethanol to each well, mixed well, and centrifuged at 3,000 g for 30 min. Two hundreds µL of 80% ethanol was used to wash again, centrifuged at 3,000 g for 5 min. After standing at room temperature for 20 min for the ethanol completely evaporated, a 15 µL formamide solution was added to dissolve the products and denatured at 95 °C for 2 min. Electrophoresis of the purified sequencing reaction products was done in an ABI 3730 sequencer (Applied Biosystem, Foster City, CA, USA).
The sequencing reaction products were electrophoresed in an ABI 3730 sequencer (Applied Biosystem, Foster City, CA, USA). The sequence data was imported into the Assign 3.5 genotype calling. HLA genotypes were assigned at the four-digit level. HLA haplotypes were analyzed using the Arlequin 3.5.1 software.
Statistical analysis
Statistical analysis was performed using SPSS (version 20.0, SPSS Inc., Chicago, IL, USA). Age and IOP were compared between patients with PSS and controls using independent-samples t-test. Hardy-Weinberg equilibrium (HWE) was evaluated using the chi-squared test. The difference in sex, allele frequency, genotype frequency, and haplotype frequency between cases and controls were evaluated using the chi-squared test or Fisher’s exact test. The maximum expectation algorithm determined haplotype frequency in the Arlequin 3.5.1 software. Multiple testing was corrected using the Bonferroni method. P<0.05 was considered statistically significant. Odds ratio (OR) and 95% confidence interval (CI) were calculated whenever applicable.
Results
Demographic and clinical features of participants
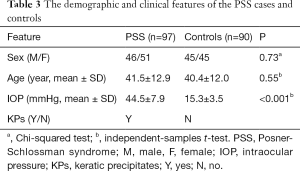
There were 46 (47.4%) males and 51 (52.6%) females in the PSS group, with an average age of 41.5±12.9 years. In the control group, there were 45 (50.0%) males and 45 (50.0%) females, with an average age of 40.4±12.0 years. No significant difference in sex and age was found between cases and controls (P=0.73 and 0.55, respectively, Table 3). The mean IOP was 44.5±7.9 mmHg in patients with PSS while 15.3±3.5 mmHg in controls. The IOP in cases was significantly higher than that in controls (P<0.001, Table 3).
Full table
HLA-E allele and genotype frequencies
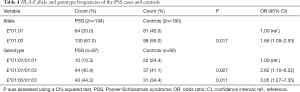
Two alleles (E*01:01 and E*01:03) and three genotypes (E*01:01/01:01, E*01:01/01:03 and E*01:03/01:03) at the HLA-E locus were detected in both PSS and control groups (Table 4). The genotype distributions of HLA-E in both groups were following HWE (P>0.11; data not shown). The allele frequency of HLA-E*01:03 in patients with PSS was significantly higher than that in the control group (P=0.017, OR =1.66, 95% CI: 1.09–2.53, Table 4), which survived the Bonferroni correction (corrected P=0.034). The genotype frequencies of HLA-E*01:01/01:03 and HLA-E*01:03/01:03 in the PSS group were significantly higher than that in the control group (P=0.027, OR =2.62, 95% CI: 1.10–6.22; P=0.011, OR =3.05, 95% CI: 1.27–7.35, respectively, Table 4).
Full table
HLA-G allele and genotype frequencies
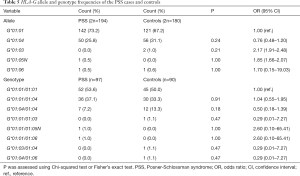
Four alleles at the HLA-G locus were detected in the PSS group (G*01:01, G*01:04, G*01:05N and G*01:06) and control group (G*01:01, G*01:03, G*01:04 and G*01:06) respectively (Table 5). Five genotypes (G*01:01/01:01, G*01:01/01:04, G*01:04/01:04, G*01:01/01:05N and G*01:01/01:06) were detected in the PSS group, while 6 genotypes (G*01:01/01:01, G*01:01/01:04, G*01:04/01:04, G*01:01/01:03, G*01:03/01:04 and G*01:04/01:06) were detected in the control group (Table 5). The top three HLA-G genotypes were G*01:01/01:01, G*01:01/01:04, and G*01:04/ 01:04. The genotype distributions of HLA-G polymorphisms in both PSS and control groups were in accordance with HWE (P>0.11; data not shown). There was no significant difference in the frequency of HLA-G alleles and genotypes between the two groups (P>0.05, Table 5).
Full table
HLA-E-G haplotype frequencies
In PSS and control groups, seven haplotypes were detected (Table 6). The top 3 HLA-E-G haplotypes were E*01:03-G*01:01, E*01:01-G*01:04, and E*01:01-G*01:01. The haplotype frequency of HLA-E*01:03-G*01:01 in the PSS group was significantly higher than that in the control group (P=0.019, OR =1.63, 95% CI: 1.08–2.46, Table 6), although this association did not survive the Bonferroni correction (corrected P=0.13). No significant difference in the other haplotypes was found between the two groups (all P>0.05, Table 6).

Full table
Discussion
HLA-E and HLA-G gene polymorphisms affect their expression levels and immune function (26-28). The HLA-E and HLA-G alleles exhibit limited polymorphisms, with only ten 4-digit HLA-E alleles and twenty-two 4-digit HLA-G alleles (IMGT/HLA, http://www.ebi.ac.uk/ipd/imgt/hla/, Release 3.36.0, 2019 April). In this study, the distributions of HLA-E and HLA-G alleles in the control group are similar to those in the normal Chinese Han population (Table 4) (21,24,29).
The frequency of the HLA-E*01:03 allele in the PSS group was significantly higher than that in the control group (Table 4), suggesting that the HLA-E*01:03 allele is a susceptibility gene for PSS. The HLA-E*01:03 allele has been associated with cytomegalovirus (CMV) infection in patients after kidney transplantation, chronic hepatitis B, rheumatoid arthritis, pemphigus vulgaris, nasopharyngeal carcinoma and ovarian cancer, while the HLA-E*01:01 allele is a protective gene for diseases such as hepatitis C virus infection, Behcet’s disease and Hodgkin’s lymphoma (16-23,25). The difference between the HLA-E*01:01 allele-encoded protein (HLA-ER) and the HLA-E*01:03 allele-encoded protein (HLA-EG) is caused by the nucleotide change from adenine to guanine (A→G) at the rs1264457 site (16,27). Compared with HLA-ER, HLA-EG is characterized by the higher expression on the surface of NK cells, higher affinity for binding to receptors, and higher stability of binding to signal peptides (26,27). Besides, several studies have shown that CMV infection may be the main pathogenic factor of PSS (3), which might inhibit the immune surveillance function through HLA-E molecules. For example, the CMV UL40 protein has a similar structure to the HLA-E signal peptide, which can up-regulate the expression of HLA-E molecules on the cell surface and enhance the binding of HLA-E molecules to inhibitory receptors (CD94/NKG2A), inhibiting the immune function of NK cells or some CTLs (30).
Polymorphisms in the coding region of the HLA-G gene can affect the coding of nucleotides. For example, the HLA-G*01:13N allele changes the first base of codon 54 (α1 domain) from cytosine to thymine (C→T), ending the coding early, resulting in the inability to synthesize the HLA-G protein (28). The HLA-G*01:05N allele deletes the last nucleotide of codon 129 or in the first nucleotide of codon 130 (exon 3), causing a stop codon in advance at the codon 189, which encodes an incomplete HLA-G protein, affecting its function (28). The HLA-G*01:04 and HLA-G*01:05N alleles are susceptibility genes for habitual abortion (23). The HLA-G*01:05N allele is also a protective gene for HIV infection, while the HLA-G*01:01:08 allele is a susceptibility gene for HIV infection (21). In this study, we did not find a significant difference in the HLA-G allele and genotype frequency between patients with PSS and controls (Table 5), indicating that the HLA-G gene polymorphisms might not be related to PSS in the southern Chinese Han population.
The HLA-E and HLA-G loci are adjacent loci at 660 kb. Studies have reported that HLA-E and HLA-G polymorphisms are simultaneously related to a variety of diseases. For instance, both the HLA-E*01:03 allele and the HLA-G 14bp INS/DEL polymorphism are associated with CMV infection (31,32). Both HLA-E*01:03 and HLA-G*01:05N alleles are protective factors for HIV-1 infection (33). The HLA-E*01:01 allele and the HLA-G*01:01:01 allele are both protective factors for Behcet’s disease (34). We found that the haplotype frequency of HLA-E*01:03-G*01:01 was higher in the PSS group than in the control group (Table 6), despite this association did not survive the Bonferroni correction. We hypothesized that the HLA-G*01:01 encoded protein as a signal peptide can enhance the stability of the HLA-E*01:03 encoded HLA-EG peptide complex and inhibited the function of NK cells or some CTLs so that the HLA-E*01:03-G*01:01 haplotype may be more likely to promote the pathogenesis of PSS. Further research must reveal the underlying mechanisms. We reported for first time that polymorphisms of non-classical HLA-Ib genes (i.e., HLA-E and HLA-G) were associated with PSS in the southern Chinese Han population. We found that the HLA-E*01:03 allele was a susceptibility gene for PSS, and the HLA-E*01:03-G*01:01 haplotype might be a risk factor for PSS. Further investigation into the expression of HLA-E and HLA-G molecules at the transcriptional and protein levels is required to must evaluate their role in the pathogenesis of PSS.
Acknowledgments
We are indebted to the participants for their excellent co-operation and support. We want to thank the volunteer blood donors for generously supplying blood samples for this study. We also thank Dr. Baojian Fan for his insightful comments on this manuscript.
Funding: This study was supported by the Science, Technology, and Innovation Commission of Shenzhen Municipality under Grant (number JCYJ20180228164400218 and GJHZ20180420180937076), Health and Family Planning Commission of Shenzhen Municipality under Grant (number SZGW2017005), and Sanming Project of Medicine in Shenzhen Grant (number SZSM201812090 and SZSM201811092).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were following the ethical standards of the local Ethics Committee of Shenzhen Eye Hospital and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. Written informed consent was obtained from all study participants.
References
- Posner A, Schlossman A. Syndrome of unilateral recurrent attacks of glaucoma with cyclitic symptoms. Arch Ophthal 1948;39:517-35. [Crossref] [PubMed]
- Jiang JH, Zhang SD, Dai ML, et al. Posner-Schlossman syndrome in Wenzhou, China: a retrospective review study. Br J Ophthalmol 2017;101:1638-42. [Crossref] [PubMed]
- Megaw R, Agarwal PK. Posner-Schlossman syndrome. Surv Ophthalmol 2017;62:277-85. [Crossref] [PubMed]
- Maruyama K, Maruyama Y, Sugita S, et al. Characteristics of cases needing advanced treatment for intractable Posner-Schlossman syndrome. BMC Ophthalmol 2017;17:45. [Crossref] [PubMed]
- Alifu M, Chang X, Kuerban G, et al. Distribution of HLA-A alleles and its relation to clinical outcome in Uyghur and Han patients with advanced squamous cell cervical cancer in Xinjiang, China. Transl Cancer Res 2018;7:220-30. [Crossref]
- Bednarczuk T, Gopinath B, Ploski R, et al. Susceptibility genes in Graves' ophthalmopathy: searching for a needle in a haystack? Clin Endocrinol (Oxf) 2007;67:3-19. [Crossref] [PubMed]
- Gil-Carrasco F, Vargas-Alarcón G, Zúñiga J, et al. HLA-DRB and HLA-DQB loci in the genetic susceptibility to develop glaucoma in Mexicans. Am J Ophthalmol 1999;128:297-300. [Crossref] [PubMed]
- Pakzad-Vaezi K, Pepple KL. Tubulointerstitial nephritis and uveitis. Curr Opin Ophthalmol 2017;28:629-35. [Crossref] [PubMed]
- Qi J, Li Q, Lin Z, et al. Higher risk of uveitis and dactylitis and older age of onset among ankylosing spondylitis patients with HLA-B*2705 than patients with HLA-B*2704 in the Chinese population. Tissue Antigens 2013;82:380-6. [Crossref] [PubMed]
- van Essen TH, Roelen DL, Williams KA, et al. Matching for Human Leukocyte Antigens (HLA) in corneal transplantation - to do or not to do. Prog Retin Eye Res 2015;46:84-110. [Crossref] [PubMed]
- Hirose S, Ohno S, Matsuda H. HLA-Bw54 and glaucomatocyclitic crisis. Arch Ophthalmol 1985;103:1837-9. [Crossref] [PubMed]
- Zhao J, Zhu T, Chen W, et al. Human Leukocyte Antigens-B and -C Loci Associated with Posner-Schlossman Syndrome in a Southern Chinese Population. PLoS One 2015;10:e0132179. [Crossref] [PubMed]
- Zhao J, Zhu T, He L, et al. Association of HLA-DPA1 and -DPB1 polymorphisms with Posner-Schlossman syndrome among southern Chinese Han population. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2015;32:254-8. [PubMed]
- Carosella ED, Gregori S, LeMaoult J. The tolerogenic interplay(s) among HLA-G, myeloid APCs, and regulatory cells. Blood 2011;118:6499-505. [Crossref] [PubMed]
- Morandi F, Pistoia V. Interactions between HLA-G and HLA-E in physiological and pathological conditions. Front Immunol 2014;5:394. [Crossref] [PubMed]
- Bhanusali DG, Sachdev A, Rahmanian A, et al. HLA-E*0103X is associated with susceptibility to Pemphigus vulgaris. Exp Dermatol 2013;22:108-12. [Crossref] [PubMed]
- Guberina H, da Silva Nardi F, Michita RT, et al. Susceptibility of HLA-E*01:03 allele carriers to develop cytomegalovirus replication after living-donor kidney transplantation. J Infect Dis 2018;217:1918-22. [Crossref] [PubMed]
- Hirankarn N, Kimkong I, Mutirangura A. HLA-E polymorphism in patients with nasopharyngeal carcinoma. Tissue Antigens 2004;64:588-92. [Crossref] [PubMed]
- Iwaszko M, Świerkot J, Kolossa K, et al. Polymorphisms within the human leucocyte antigen-E gene and their associations with susceptibility to rheumatoid arthritis as well as clinical outcome of anti-tumour necrosis factor therapy. Clin Exp Immunol 2015;182:270-7. [Crossref] [PubMed]
- Martín P, Krsnik I, Navarro B, et al. HLA allele E*01: 01 is associated with a reduced risk of EBV-related classical Hodgkin Lymphoma independently of HLA-A*01/*02. PLoS One 2015;10:e0135512. [Crossref] [PubMed]
- Matte C, Lajoie J, Lacaille J, et al. Functionally active HLA-G polymorphisms are associated with the risk of heterosexual HIV-1 infection in African women. AIDS 2004;18:427-31. [Crossref] [PubMed]
- Schulte D, Vogel M, Langhans B, et al. The HLA-E(R)/HLA-E(R) genotype affects the natural course of hepatitis C virus (HCV) infection and is associated with HLA-E-restricted recognition of an HCV-derived peptide by interferon-gamma-secreting human CD8(+) T cells. J Infect Dis 2009;200:1397-401. [Crossref] [PubMed]
- Yan WH, Fan LA, Yang JQ, et al. HLA-G polymorphism in a Chinese Han population with recurrent spontaneous abortion. Int J Immunogenet 2006;33:55-8. [Crossref] [PubMed]
- Zheng H, Lu R, Xie S, et al. Human leukocyte antigen-E alleles and expression in patients with serous ovarian cancer. Cancer Sci 2015;106:522-8. [Crossref] [PubMed]
- Zidi I, Laaribi AB, Bortolotti D, et al. HLA-E polymorphism and soluble HLA-E plasma levels in chronic hepatitis B patients. HLA 2016;87:153-9. [Crossref] [PubMed]
- Kaiser BK, Pizarro JC, Kerns J, et al. Structural basis for NKG2A/CD94 recognition of HLA-E. Proc Natl Acad Sci 2008;105:6696-701. [Crossref] [PubMed]
- Gonçalo M. HLA-B*58:01 is not the only risk factor associated with allopurinol-induced severe cutaneous adverse drug reactions. Ann Transl Med 2018;6:S7. [Crossref] [PubMed]
- Castelli EC, Veiga-Castelli LC, Yaghi L, et al. Transcriptional and posttranscriptional regulations of the HLA-G gene. J Immunol Res 2014;2014:734068. [PubMed]
- Wang WY, Tian W, Liu XX, et al. HLA-G coding region and 3'untranslated region (3'UTR) in two Chinese Han populations. Immunol Lett 2016;176:65-71. [Crossref] [PubMed]
- Prod'homme V, Tomasec P, Cunningham C, et al. Human cytomegalovirus UL40 signal peptide regulates cell surface expression of the NK cell ligands HLA-E and gpUL18. J Immunol 2012;188:2794-804. [Crossref] [PubMed]
- Romagnani C, Pietra G, Falco M, et al. HLA-E-restricted recognition of human cytomegalovirus by a subset of cytolytic T lymphocytes. Hum Immunol 2004;65:437-45. [Crossref] [PubMed]
- Zheng XQ, Zhu F, Shi WW, et al. The HLA-G 14 bp insertion/deletion polymorphism is a putative susceptible factor for active human cytomegalovirus infection in children. Tissue Antigens 2009;74:317-21. [Crossref] [PubMed]
- Lajoie J, Hargrove J, Zijenah LS, et al. Genetic variants in nonclassical major histocompatibility complex class I human leukocyte antigen (HLA)-E and HLA-G molecules are associated with susceptibility to heterosexual acquisition of HIV-1. J Infect Dis 2006;193:298-301. [Crossref] [PubMed]
- Park KS, Park JS, Nam JH, et al. HLA-E*0101 and HLA-G*010101 reduce the risk of Behcet's disease. Tissue Antigens 2007;69:139-44. [Crossref] [PubMed]


