
Use of radiomic features and support vector machine to distinguish Parkinson’s disease cases from normal controls
Introduction
Parkinson’s disease (PD) is a common chronic progressive neurodegenerative condition in the elderly caused by early death of dopaminergic neurons in the central nervous system (1,2). The destruction of dopaminergic neurons in PD patients produces a variety of motor and non-motor symptoms. Typical motor symptoms include bradykinesia, muscle stiffness, and resting tremor. Major non-motor symptoms include depression, psychosis, falls, genitourinary diseases, and sleep disorders (3-5). These motor and non-motor symptoms seriously affect the quality of life of patients with PD (5). Moreover, the prevalence of PD is increasing year by year as the population ages, and is expected to cause huge economic and social burden to society (6). Therefore, accurate early diagnosis of PD is essential for timely treatment and intervention.
Some studies have shown that clinical non-motor symptoms and biomarkers such as cerebrospinal fluid (CSF) have been used as indicators for the early diagnosis of PD (7-9). Although these indicators have outstanding diagnostic capabilities, clinical application is often challenging because of the invasive nature and costs incurred. Currently, medical imaging technology mainly including magnetic resonance imaging (MRI), positron emission tomography (PET), and single photon emission computed tomography (SPECT) has drawn attention as an effective tool for early diagnosis of PD (10). PET is commonly used for imaging to measure brain dysfunction (11). In particular, the use of 18F-fluorodeoxyglucose PET (18F-FDG PET) imaging to measure glucose consumption allows us to identify specific brain metabolic patterns in early-stage neurodegenerative disease (12). Previous studies have reported regional differences and proposed several imaging markers in brain glucose metabolism in PD (13-18). In summary, the development of medical imaging technology has made imaging markers a widely used clinical indicator for early diagnosis of PD and promoted the rapid development of computer-aided diagnosis.
At present, with the development of machine learning and deep learning, increasing computer-aided diagnostic categories are used to identify PD or other forms of Parkinson’s syndrome based on different brain imaging modalities (19-21). Computer-aided PD diagnosis has the advantages of accurate quantification and reproducibility. It can help doctors and patients avoid unnecessary medical examinations, potential side effects, and safety risks (22). For example, Juh et al. (23) used statistical parametric mapping to determine useful metabolic markers in diagnosing PD. Eckert et al. (24) combined visual assessment of individual scans with blinded computer assessment in the differential diagnosis of PD. Shen et al. (25) improved a framework based on Group Lasso Sparse Deep Belief Network (GLS-DBN) to distinguish between PD and normal controls (NC) subjects based on FDG-PET imaging, and established the computer-aided classifier for PD and NC.
However, most of the markers used in these methods are low-level features that do not accurately reflect the neuropathological heterogeneity of brain tissue associated with PD, and thereby do not provide precise diagnostic results. In contrast, although deep features from deep learning methods can reflect good diagnostic capabilities, it is difficult to explain the clinical correlation between these deep features and the actual disease. Therefore, high-order feature extraction methods are required.
Radiomic approach could be an alternative method to solve above problems. Radiomic approach has ensured the conversion of medical images into mineable, high-dimensional data and can quickly extract a large number of high-level advanced features from medical images by using high-throughput calculations (26-29). These radiomic features reflect the potential pathophysiological information and contribute to clinical diagnosis (30). Although widely used in oncology research (31-34), it has recently been extended to other medical applications such as neurodegenerative diseases. Some studies have used it to diagnose Alzheimer’s disease (AD) and mild cognitive impairment (MCI) (35-37). Therefore, we hypothesized that radiomic features may describe the brain tissue heterogeneity in PD to effectively distinguish PD patients from NC.
The present study was designed to determine whether radiomic features extracted from 18F-FDG PET brain images can be used for computer-assisted diagnosis of PD and NC. We also proposed a new diagnostic method for computer-assisted PD based on radiomics.
Methods
Figure 1 shows the framework of this study. First, the image was spatially normalized and smoothed. Then, the concerned brain regions were determined based on atlas-based method from literatures (38,39). Subsequently, the radiomic features were extracted from the brain regions of interest (ROIs). Thereafter, we used autocorrelation and fisher score algorithms to deduce the correlation between features and to select the most efficient radiomic features for classification. Finally, we completed the classification for PD vs. NC using support vector machine (SVM). The details of this approach are described in subsequent sections.
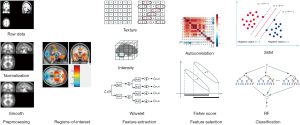
Materials
In this retrospective multicohort study, the data used were derived from two cohorts: (I) 182 subjects from Huashan Hospital (Fudan University, Shanghai, China) comprising 91 NC and 91 PD patients. (II) 48 subjects from Wuxi 904 Hospital (Jiangsu, China) comprising 26 NC and 22 PD patients.
In the case of PD patient cohorts, all subjects were screened and clinically examined by two senior investigators of movement disorders prior to being included in the study. Based on the UK Brain Bank criteria (40), a confirmed diagnosis of PD was made in all subjects if the patients had “pure” parkinsonism without a history of known causative factors such as encephalitis or neuroleptic treatment, and did not have dementia, supranuclear gaze abnormalities, or ataxia. Unified Parkinson’s Disease Rating Scale (UPDRS) motor examination (items 18–31) and Hoehn & Yahr scale (H&Y) were assessed at least 12 h after the cessation of oral antiparkinsonian medications (i.e., a practically defined “off” condition) and not more than 2 h before the PET scan.
As for NC subjects, all subjects underwent the same clinical screening procedures as PD patients and showed negatively in these tests. The following exclusion criteria were applied: (I) a history of neurological or psychiatric illness; (II) prior exposure to neuroleptic agents or drug use; (III) an abnormal neurological examination. To control the demographic differences among patients and controls, 91 age- and gender-matched NC (Huashan cohort) together with 26 age- and gender-matched NC (Wuxi cohort) were enrolled in our study.
Image acquisition and preprocessing
Before PET scanning, all participants were required to fast for at least 6 h but had free access to water. PET scans were performed with a Siemens Biograph 64 HD PET/computed tomography (CT) (Siemens, Germany) in 3D mode. Prior to the emission scan, a low-dose CT transmission scan was performed for attenuation correction. The brain emission scan was acquired between 45 and 55 min after intravenous injection of 185 MBq of 18F-FDG. Data was reconstructed using 3D ordered subset expectation maximization and corrected for random coincidences, scatter, and radioactive decay. As no arterial blood sampling was performed in this clinical imaging protocol, we used radioactivity count images to measure changes in relative regional glucose metabolism. All studies in PD patients and NC were carried out under standardized circumstances, i.e., in a quiet and dimly lit room with minimal background noise and in a resting state with the eyes open.
For each subject, the scanned PET image was first spatially normalized to the Montreal Neurological Institute (MNI) space (FDG-PET template) with linear and non-linear 3D transformations, which made the image size and resolution consistent. Next, the normalized PET images were smoothed using a Gaussian smoothing kernel with a full-width at half maximum (FWHM) value of 10×10×10 mm3 that could blur image edges and improve the signal-to-noise ratio. As a result, the preprocessed images had a spatial resolution of 79×95×69 with a voxel size of 2×2×2 mm3.
Image data were preprocessed using Statistical Parametric Mapping 12 (SPM12, Wellcome Department of Imaging Neuroscience, Institute of Neurology, London, UK, http://www.fil.ion.ucl.ac.uk/spm/) software implemented in MATLAB R2018a (Mathworks Inc, Sherborn, MA, USA).
Definition of regions-of-interest
According to various previous studies, the morphological and metabolic changes in some brain regions were directly related to the pathology of PD. Metabolic changes in relevant brain regions of PD patients have been detected using 18F-FDG PET (18,41). Therefore, we referred to previous studies to select brain regions with morphological and metabolic changes, including the superior frontal (24,41,42), the middle frontal (24), supplementary motor area (43), occipital gyrus (24), caudate nucleus (24,44), putamen (42), pallidum (41), thalamus (24,41,42,44), inferior temporal (24,44), cerebellum (24,41,42), and pons (45).
Figure 2 shows all the brain ROIs (n=95). We selected 90 regions from the anatomical automatic labeling (AAL) template (46), and 5 other regions where functional imaging studies in PD commonly report altered metabolism, including bilateral cerebellum, bilateral pons and cerebellar vermis.
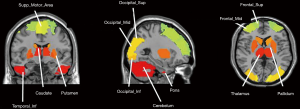
To verify the effectiveness of above brain ROIs, we further used a two-sample Student’s t-test in SPM12 for group comparisons in Huashan cohort. In this step we set the peak threshold to P<0.001 and performed family wise error (FWE) correction throughout the brain region (35,47).
Radiomic features extraction
In this section, we extracted features by using the radiomics tool developed by Vallieres et al. (https://github.com/mvallieres/radiomics). We used “Texture Toolbox” in the radiomics tool to perform texture analysis from each input ROI. All steps were performed in MATLAB R2018a, including wavelet band-pass filtering, isotropic resampling, Lloyd-Max quantization, and feature calculation. Each 18F-FDG PET image was prepared for intensity analysis, matrix-based texture analysis, and wavelet analysis at a scale of 2 mm. Firstly, the wavelet band-pass filtering was carried out by applying different weights to bandpass sub-bands (LHL, LHH, LLH, HLL, HHL, and HLH) of the volume of interest (VOI), compared to low- and high-frequency sub-bands (LLL and HHH) in the wavelet domain. The ratio of the weight was defined by R, and the values of R were 1/2, 2/3, 1 (no wavelet filtering), 3/2, and 2. Secondly, isotropic resampling was performed at the initial in-plane resolution of every PET image, the Lloyd-Max quantization algorithm was applied to normalize the PET images to 256 gray-level images. Finally, we obtained four types of texture matrices [gray-level co-occurrence matrix (GLCM), gray-level run-length matrix (GLRLM), gray-level size zone matrix (GLSZM), and neighborhood gray-tone difference matrix (NGTDM)] from quantized PET images. Based on above texture matrices, we achieved 43 texture features, 188 wavelet features and 4 intensity features. 43 texture features include 3 histogram-based textures, 9 texture features from the GLCM, 13 texture features from the GLRLM, 13 texture features from the GLSZM, 5 texture features from the NGTDM. Four intensity features include SUVmax, SUVpeak, SUVmean, aucCSH. Detailed radiomic features are listed in Table 1.

Full table
Features selection
In order to reduce the number of features, we ensured independence among features and only selected those features that make an important contribution for the classification step. In this step, the clinical basic information (age and gender) was also added into the radiomic features. A 5-fold cross-validation algorithm was carried out in this procedure.
The feature selection step was performed through two steps: feature autocorrelation and fisher score algorithm. First, feature autocorrelation was performed to reduce redundancy between high-dimensional features. For each feature, the average absolute correlation based on pair-wise correlations was calculated, as defined by the following formula:

If in a pair-wise correlation, the Ci,j value was greater than 0.8, the feature with the higher average absolute correlation was removed.
Next, we used fisher score algorithm to rank the remaining features. Fisher score algorithm is a filter-based method for supervised feature selection. It selects each feature independently according to their scores under the fisher criterion. After computing the fisher score for each feature, the top-m ranked features with large scores could be retained. In this study, we retained the top-30 ranked features. These selected features were used for subsequent classification. Finally, we repeated the above steps in 500 times. All the above-described feature selection steps were implemented in MATLAB R2018a.
SVM classification
To verify the diagnostic capabilities of the selected radiomic features, we used SVM classifiers for classification experiments. The feature normalization was first performed before classification. We used the Min-Max Normalization method on the training and test sets to normalize the selected radiomic features. Then, we performed an NC vs. PD classification using SVM classifier. The SVM is a supervised learning method that has been widely used in statistical classification and regression analysis. It maps the vector to a higher dimensional space in which a maximum interval hyperplane is created. Therefore, these vectors from the higher dimensional space could achieve high classifying performance. In this study, three kernel (linear, sigmoid, and radial basis) functions were used to detect feature generalization ability and classification reliability. The Wuxi cohort was used as an additional test dataset (test 2) to test the model. We calculated the mean [± standard deviation (SD)] accuracy, sensitivity, and specificity of the 500 repetitions as the final result. In addition, we used the receiver operating characteristic (ROC) graph to visualize the experimental results with the test dataset. The area under the curve (AUC) of the ROC was also computed to quantitatively evaluate the classification performance.
In addition, to further validate the classifying efficiency of the radiomic features, we applied another classifier: “random forest (RF)”. 30 was set as the number of trees (54,55).
Comparative experiment
To verify the superiority of high-level radiomic features in selected brain regions, we deployed the following two comparative experiments.
For the first comparative experiment, we calculated the average voxel value based on each ROI in the brain and obtained 26 average voxel values for each subject. We considered these 26 values as features for the classification of NC and PD. For the second comparative experiment, we combined the radiomic features and the average voxel value of each ROI in the brain as features and performed a classification of NC and PD by the same classifier as above.
In the two comparative experiments, the 5-fold cross-validation was performed in Huashan cohort, in which 80% data was used as the training set and the remaining 20% data was used as the first test set (test 1). The Wuxi cohort was used as the second test set (test 2). The comparative experiments underwent the same process procedures with 500 repetitions.
Correlation between selected features and clinical scales
To verify the reproducibility of selected radiomic features and their relevance to clinical scales, we selected features with more frequent occurrences and used these features to verify the correlations between them and the clinical scales. We calculated the Pearson correlation coefficient between these features and UPDRS and Spearman correlation coefficient between these features and H&Y.
Furthermore, we calculated the Pearson correlation coefficient between the top three high-frequency features and the average voxel value of the related brain region where they were located in the two test sets.
Decision score
To better describe the discrimination ability of the results, a decision score could be output after the SVM model decision analysis to represent the model score of NC or PD; we used this score to perform the t-test on NC and PD to observe the intergroup differences. In the experiment, we calculated the average scores of NC and PD in the test set (test 2).
Statistical analysis
The clinical and demographic characteristics (age, sex, H&Y, UPDRS) were assessed using two-sample t-test and chi-square test. Morphological differences in brain regions in NC and PD patients were verified using the two-sample two-tailed Student’s t-test on all NC and PD patients from the Huashan cohort. High-frequency features associated with clinical scales and the average voxel values were calculated using Pearson correlation coefficients. All statistical analyses were performed using MATLAB R2018a run on a Windows platform. Values were considered significant for P<0.05.
Results
Subjects
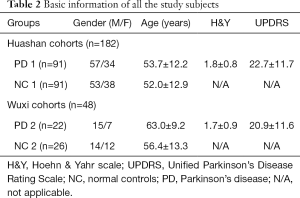
The subject demographics and clinical characteristics for the two diagnostic groups in this study are detailed in Table 2. There were no significant differences in age and sex among the NC and PD patients (all P>0.05; χ2 test). In addition, the H&Y and UPDRS scores of the Huashan cohort were relatively slightly higher than the Wuxi cohort.
Full table
Brain ROIs validation
Brain regions from the Huashan cohort are shown in Table 3. The results were basically consistent with the selected brain regions based on the atlas-based method. The brain regions after t-test were the frontal lobe regions (Frontal_Mid, Frontal_Sup, Frontal_Inf_Orb); temporal lobe regions (Temporal_Mid, Temporal_Inf); occipital lobe regions (Occipital_Mid, Occipital_Sup, Occipital_Inf); and cerebellum region (Cerebellum).

Full table
Radiomic features extraction and selection
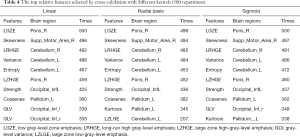
After feature extraction, we got a total of 6,110 features. After the autocorrelation step, 900–1,000 types of uncorrelated features remained. After the fish score test, we retained the top-30 ranked features. Table 4 lists the 10 most frequent features, their occurrence times, and the brain areas of their locations in the 500 repetitions performed during cross-validation with three different kernels. In general, these features showed excellent consistency in repeated experiments and they have good pathological revealing ability.
Full table
Classification performance
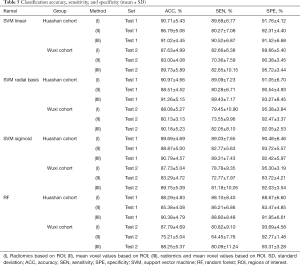
As shown in Table 5, using the selected radiomic features with radial basis kernel in the Huashan cohort, the average accuracy, sensitivity, and specificity of the test set to distinguish NC from PD patients was 90.97%±4.66%, 89.09%±7.23%, and 91.05%±6.70%, respectively. The classification performance of linear and sigmoid kernels was slightly poor, with average accuracies of 90.71%±5.43% and 89.69%±4.69%, respectively. In the results of the first comparison experiment, the average accuracies of the three kernels based on the average voxel value within the Huashan cohort test dataset were 86.79%±5.06%, 88.51%±4.52%, and 88.87%±5.00% respectively. Additionally, in the Wuxi test dataset, the classification results based on our proposed method were 87.63%±4.99%, 88.08%±5.27%, and 87.73%±5.04%, which was better than the traditional voxel values method (83.00%±4.08%, 80.13%±3.13%, and 83.29%±4.72% respectively). Furthermore, when we combined the radiomic features and the voxel values, higher classification accuracies could be achieved. The highest average accuracy, sensitivity, and specificity in the Huashan cohort were 91.26%±5.15%, 89.43%±7.17%, and 93.27%±8.45%. In the Wuxi test dataset, they were 90.18%±5.23%, 82.05%±8.10%, and 92.05%±2.53%, respectively. That means that the radiomic features provided additional classification information for the traditional voxel values information.
Full table
Figure 3 presents the different kernel functions (linear, radial basis, sigmoid) for the ROC curve in the classification experiment. The average AUCs (± SD) of the ROC curve of the linear kernel function were 0.82±0.02, 0.81±0.03, and 0.87±0.03 (Figure 3A). The average AUCs (± SD) of the ROC curve of the radial basis kernel function were 0.85±0.03, 0.78±0.03, and 0.88±0.04 (Figure 3B). The average AUCs (± SD) of the ROC curve of the sigmoid kernel function were 0.85±0.03, 0.80±0.02, and 0.87±0.02 (Figure 3C).

In the classification experiments with the RF classifier, we also observed that the radiomic features achieved higher classification accuracies than the traditional voxel values method in both Huashan (88.29%±4.83% vs. 85.38%±4.08%) and Wuxi cohorts (87.79%±4.69% vs. 75.21%±5.04%). The combined model achieved the highest classification accuracies, which was consistent with the results using SVM classifiers.
Correlation analysis
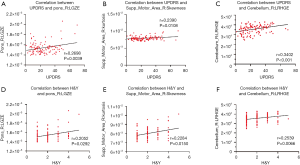
As shown in Figure 4, the high-frequency features, including low gray-level zone emphasis (LGZE), long-run high gray-level emphasis (LRHGE), and skewness, were all significantly correlated with clinical scale values in the test dataset. LGZE in the right of the pons was significantly correlated with UPDRS (r=0.2698, P=0.0039) (Figure 4A), and H&Y (r=0.2052, P=0.0292) (Figure 4B). The features skewness and LRHGE were also both positively correlated with UPDRS (r=0.2390, P=0.0108 and r=0.3402, P<0.001, respectively) (Figure 4C,E), and H&Y (r=0.2284, P=0.0150 and r=0.2539, P=0.0066, respectively) (Figure 4D,F).
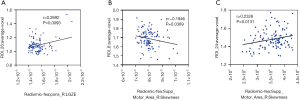
As seen in Figure 5, there was a correlation between high-frequency radiomic features and the average voxel value of its brain region. We could see that there was a negative correlation between skewness and the average voxel value of the right supper motor area (r=–0.1946, P=0.0389) (Figure 5B). The feature LGZE was positively correlated with the average voxel value of the right pons (r=0.2692, P=0.0093) (Figure 5A). The feature LRHGE was also positively correlated with the average voxel value of the right cerebellum (r=0.2328, P=0.0131) (Figure 5C).
Decision score
In our proposed method, the performance of the SVM output decision scores with different kernels used to classify NC and PD in test 2 are shown in Figure 6. Decision scores of HC were significantly higher than that of PD (linear: 2.29±1.29 vs. –1.09±1.06, P<0.001 (Figure 6A); radial basis:1.15±0.58 vs. –0.48±0.47, P<0.001 (Figure 6B); sigmoid: 1.20±0.72 vs. –0.45±0.49, P<0.001) (Figure 6C). Furthermore, the SVM with different kernel output decision scores could be used as a quantitative imaging biomarker for classification, with significant differences between the NC and PD groups.

Discussion
In this study, we employed statistical analysis and two kinds of classifier to implement a 18F-FDG-PET imaging-based radiomic method for the diagnosis of PD. To demonstrate the stability and applicability of our proposed radiomic methods, we selected different samples from different PET scanners with different imaging characteristics, including cohorts from Huashan Hospital and Wuxi 904 Hospital. This cross-sample research method could test the generalization ability of the model (26). Overall, this study suggested that radiomic features can be used for early computer-aided diagnosis of PD.
Additionally, in feature selection, we listed 10 features with frequent occurrences. Among them, we found that “LGZE”, “skewness” and “LRHGE” appeared multiple times in different brain areas, indicating that these features have good ability to indicate pathology. LGZE is a textural measure (56). One 18F-FDG-PET study in follicular lymphoma found that LGZE could solely predict complete response of chemotherapy (57). Yet further study is needed to help understand its meaning in PD. Skewness is a textural measure of asymmetry (58). It has scarcely been investigated in the field of neurological PET (59). A recent study analyzed textural parameters extracted from amyloid PET in AD and found that histogram-based indices [skewness (15%) and cumulative histogram (AUC: 35%)] were highly correlated with the subject’s status and hence strong markers of AD progression, as they reflected the prevalence of high-intensity and low-intensity voxels in positive and negative scans, respectively (60). Thus, combined with our findings, we inferred that the “skewness” feature we found in PD by 18F-FDG-PET might also reflect the specific status of PD and be used as a potential biomarker. The LRHGE feature is expected to be large for images with many long runs and high gray-level values (50). This feature was mainly found in oncology research (61-64). For example, a study on breast cancer showed that LRHGE was associated with risk categories as well as the neoadjuvant chemotherapy response (61). Another study on cervical cancer found that LRHGE was correlated with the differentiation (62). To our best knowledge, this is the first mention of the LRHGE to feature in the field of neuropathy. Further studies are required to understand whether LRHGE could help to distinguish between the different subtypes of PD or offer other important information.
In clinics, there is a disease-specific anatomical pattern in PD which is characterized by relatively increased metabolism in the Globus pallidus and putamen, thalamus, cerebellum, pons and sensorimotor cortex, and relative decreased metabolism in the lateral frontal and parietooccipital areas (18,65-67). The brain regions involved in the top relative features selected in this study were consistent with this anatomical pattern. In particular, we observed that high-frequency features from this study were mainly located in the pons, supper motor area and cerebellum. As PD is mainly a movement disorder, it was reasonable to find these disease specific features in pons and the super motor area (68-72). The cerebellum plays an important role in the pathophysiology of PD, both in terms of symptomology and compensation for the damaged and dysfunctional striato-thalamo-cortical pathway (64). Indeed, cerebellar dysfunction is considered as the common link between essential tremor and resting tremor in PD (68). A functional MRI study (69) also identified the role of cerebellar circuitry in PD and underscored the involvement of the cerebello-thalamo-cortical pathways in tremorgenesis. Similar previous research supports the correlations between the features of cerebellum and clinical scales reported in this study. In addition, many studies (70-73) have suggested that cerebellar activity and changes of functional and effective connectivity within the cerebello-thalamo-cortical pathway activity are possible compensatory mechanisms in PD. Thus, we believed that the high-frequency features we found in the cerebellum of PD patients might help to further explore and understand such mechanisms.
As shown in Figure 4, the correlation of statistical results verified that clinical scale is indeed relevant to these high-frequency radiomic features. Moreover, the correlation between these features and the voxel values in the brain region suggested that they may be able to replace traditional voxel values in clinical PD diagnosis. As voxel-based studies have already shown the presence of disease-related metabolic characteristics in PD, the correlation between them showed that our radiomic features could reflect the information provided by voxel-analysis to an extent. Furthermore, the listed brain regions indicate that these regions are significantly different in NC and PD patients. As expected, the radiomic features performed better in the classification than the traditional average voxel values in diagnosis of PD.
In addition, we also compared the classification results between our proposed method and the deep learning method. The results are shown in Table 6. With the same training and test datasets, Shen et al. (25) proposed a framework based on GLS-DBN to distinguish between PD and NC subjects. The classification accuracy achieved 90.0% in that study. This comparative result means that our proposed radiomic approach achieve a similar result with the deep learning method.

Full table
Although our proposed method showed good performance in assisting PD diagnosis, it also has some limitations. First, our work focused on PET features only. Multimodal data such as MRI and other imaging features can also be used for classification in the future. Second, the pathobiological mechanisms of the correlation between radiomic features and clinical scales were not explored. In our study, we only selected radiomic features and clinical scales to select radiomic features for classification, indicating that there was indeed a correlation between radiomic features and PD pathology; however, we did not further explore the pathobiological mechanisms. In future research, the mechanism of each feature and disease should be studied in detail. Finally, our study was based only on PD and NC samples, and it would be equally meaningful to further distinguish between different forms of Parkinson’s syndrome in the future.
Conclusions
This study proved that high-order radiomic features extracted from 18F-FDG PET brain images can be used for computer-aided diagnosis of PD, as the radiomic features for diagnosis have higher accuracy than traditional voxel values. Future research can use high-order radiomic features as quantitative biomarkers for early diagnosis of PD, such that patients may benefit from timely intervention.
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China (grant numbers 61603236, 81671239, 81361120393, 81401135, 81771483, and 81361120393), the National Key Research and Development Program of China (grant numbers 2016YFC1306305, 2016YFC1306500 and 2018YFC1707704) from Ministry of Science and Technology of China, Shanghai Technology and Science Key Project in Healthcare (grant number 17441902100), Science and Technology Commission of Shanghai Municipality (grant number 17JC1401600), Shanghai Municipal Science and Technology Major Project (grant number 2017SHZDZX01) and the Open Project Funding of Human Phenome Institute (grant number HUPIKF2018203), Fudan University.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted on the recommendation of the Ethics Committee of Huashan Hospital Affiliated to Fudan University, and written informed consent was obtained from all subjects. The protocol was approved by Huashan Hospital Affiliated to Fudan University (ID: KY 2013-336).
References
- Rocha EM, De Miranda B, Sanders LH. Alpha-synuclein: pathology, mitochondrial dysfunction and neuroinflammation in Parkinson’s disease. Neurobiol Dis 2018;109:249-57. [Crossref] [PubMed]
- Rahmim A, Huang P, Shenkov N, et al. Improved prediction of outcome in Parkinson’s disease using radiomics analysis of longitudinal DAT SPECT images. Neuroimage Clin 2017;16:539-44. [Crossref] [PubMed]
- Hussl A, Seppi K, Poewe W. Nonmotor symptoms in Parkinson’s disease. Expert Rev Neurother 2013;13:581-3. [Crossref] [PubMed]
- Chaudhuri KR, Healy DG, Schapira AH, et al. Non-motor symptoms of Parkinson’s disease: diagnosis and management. Lancet Neurol 2006;5:235-45. [Crossref] [PubMed]
- Müller B, Assmus J, Herlofson K, et al. Importance of motor vs. non-motor symptoms for health-related quality of life in early Parkinson’s disease. Parkinsonism Relat Disord 2013;19:1027-32. [Crossref] [PubMed]
- de Lau LM, Breteler MM. Epidemiology of Parkinson’s disease. Lancet Neurol 2006;5:525-35. [Crossref] [PubMed]
- Cernak M, Orozco-Arroyave JR, Rudzicz F, et al. Characterisation of voice quality of Parkinson’s disease using differential phonological posterior features. Comput Speech Lang 2017;46:196-208. [Crossref]
- Shi M, Bradner J, Hancock AM, et al. Cerebrospinal fluid biomarkers for Parkinson disease diagnosis and progression. Ann Neurol 2011;69:570-80. [Crossref] [PubMed]
- Bhat S, Acharya UR, Hagiwara Y, et al. Parkinson’s disease: cause factors, measurable indicators, and early diagnosis. Comput Biol Med 2018;102:234-41. [Crossref] [PubMed]
- Politis M. Neuroimaging in Parkinson disease: from research setting to clinical practice. Nat Rev Neurol 2014;10:708-22. [Crossref] [PubMed]
- Zhu L, Ploessl K, Kung HF. PET/SPECT imaging agents for neurodegenerative diseases. Chem Soc Rev 2014;43:6683-91. [Crossref] [PubMed]
- Teune LK, Bartels AL, Leenders KL. FDG-PET imaging in neurodegenerative brain diseases. In: Signorelli F, Chirchiglia D. editors. Functional brain mapping and the endeavor to understand the working brain. Rijeka: InTech 2013. doi: . [Crossref]
- Michell AW, Lewis SJ, Foltynie T, et al. Biomarkers and Parkinson’s disease. Brain 2004;127:1693-705. [Crossref] [PubMed]
- Saeed U, Compagnone J, Aviv RI, et al. Imaging biomarkers in Parkinson’s disease and Parkinsonian syndromes: current and emerging concepts. Transl Neurodegener 2017;6:8. [Crossref] [PubMed]
- Brooks DJ, Pavese N. Imaging biomarkers in Parkinson’s disease. Prog Neurobiol 2011;95:614-28. [Crossref] [PubMed]
- Ma Y, Tang C, Spetsieris PG, et al. Abnormal metabolic network activity in Parkinson’s disease: test-retest reproducibility. J Cereb Blood Flow Metab 2007;27:597-605. [Crossref] [PubMed]
- Tomše P, Jensterle L, Rep S, et al. The effect of 18F-FDG-PET image reconstruction algorithms on the expression of characteristic metabolic brain network in Parkinson’s disease. Phys Med 2017;41:129-35. [Crossref] [PubMed]
- Wu P, Wang J, Peng S, et al. Metabolic brain network in the Chinese patients with Parkinson’s disease based on 18F-FDG PET imaging. Parkinsonism Relat Disord 2013;19:622-7. [Crossref] [PubMed]
- Brooks DJ. PET studies on the early and differential diagnosis of Parkinson’s disease. Neurology 1993;43:S6-16. [PubMed]
- Hirschauer TJ, Adeli H, Buford JA. Computer-aided diagnosis of Parkinson’s disease using enhanced probabilistic neural network. J Med Syst 2015;39:179. [Crossref] [PubMed]
- Amoroso N, La Rocca M, Monaco A, et al. Complex networks reveal early MRI markers of Parkinson’s disease. Med Image Anal 2018;48:12-24. [Crossref] [PubMed]
- Adeli E, Shi F, An L, et al. Joint feature-sample selection and robust diagnosis of Parkinson’s disease from MRI data. Neuroimage 2016;141:206-19. [Crossref] [PubMed]
- Juh R, Kim J, Moon D, et al. Different metabolic patterns analysis of Parkinsonism on the 18F-FDG PET. Eur J Radiol 2004;51:223-33. [Crossref] [PubMed]
- Eckert T, Barnes A, Dhawan V, et al. FDG PET in the differential diagnosis of parkinsonian disorders. Neuroimage 2005;26:912-21. [Crossref] [PubMed]
- Shen T, Jiang J, Lin W, et al. Use of overlapping group LASSO sparse deep belief network to discriminate Parkinson’s disease and normal control. Front Neurosci 2019;13:396. [Crossref] [PubMed]
- Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016;278:563-77. [Crossref] [PubMed]
- Kumar V, Gu Y, Basu S, et al. Radiomics: the process and the challenges. Magn Reson Imaging 2012;30:1234-48. [Crossref] [PubMed]
- Lambin P, Leijenaar RTH, Deist TM, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017;14:749-62. [Crossref] [PubMed]
- Lambin P, Rios-Velazquez E, Leijenaar R, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 2012;48:441-6. [Crossref] [PubMed]
- Panth KM, Leijenaar RT, Carvalho S, et al. Is there a causal relationship between genetic changes and radiomics-based image features? An in vivo preclinical experiment with doxycycline inducible GADD34 tumor cells. Radiother Oncol 2015;116:462-6. [Crossref] [PubMed]
- Zhou H, Vallières M, Bai HX, et al. MRI features predict survival and molecular markers in diffuse lower-grade gliomas. Neuro Oncol 2017;19:862-70. [Crossref] [PubMed]
- Vallières M, Kay-Rivest E, Perrin LJ, et al. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci Rep 2017;7:10117. [Crossref] [PubMed]
- Li Y, Liu X, Qian Z, et al. Genotype prediction of ATRX mutation in lower-grade gliomas using an MRI radiomics signature. Eur Radiol 2018;28:2960-8. [Crossref] [PubMed]
- Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 2014;5:4006. [Crossref] [PubMed]
- Li Y, Jiang J, Lu J, et al. Radiomics: a novel feature extraction method for brain neuron degeneration disease using 18F-FDG PET imaging and its implementation for Alzheimer’s disease and mild cognitive impairment. Ther Adv Neurol Disord 2019;12:1756286419838682. [Crossref] [PubMed]
- Feng F, Wang P, Zhao K, et al. Radiomic features of hippocampal subregions in Alzheimer’s disease and amnestic mild cognitive impairment. Front Aging Neurosci 2018;10:290. [Crossref] [PubMed]
- Martínez G, Vernooij RW, Fuentes Padilla P, et al. 18F PET with flutemetamol for the early diagnosis of Alzheimer’s disease dementia and other dementias in people with mild cognitive impairment (MCI). Cochrane Database Syst Rev 2017;11:CD012884. [PubMed]
- Pfefferbaum A, Rohlfing T, Rosenbloom MJ, et al. Combining atlas-based parcellation of regional brain data acquired across scanners at 1.5 T and 3.0 T field strengths. Neuroimage 2012;60:940-51. [Crossref] [PubMed]
- Aribisala BS, Cox SR, Ferguson KJ, et al. Assessing the performance of atlas-based prefrontal brain parcellation in an aging cohort. J Comput Assist Tomogr 2013;37:257-64. [Crossref] [PubMed]
- Gibb WR, Lees AJ. The relevance of the Lewy body to the pathogenesis of idiopathic Parkinson’s disease. J Neurol Neurosurg Psychiatry 1988;51:745-52. [Crossref] [PubMed]
- Garg A, Appel-Cresswell S, Popuri K, et al. Morphological alterations in the caudate, putamen, pallidum, and thalamus in Parkinson’s disease. Front Neurosci 2015;9:101. [Crossref] [PubMed]
- Eckert T, Tang C, Eidelberg D. Assessment of the progression of Parkinson’s disease: a metabolic network approach. Lancet Neurol 2007;6:926-32. [Crossref] [PubMed]
- Yu R, Liu B, Wang L, et al. Enhanced functional connectivity between putamen and supplementary motor area in Parkinson’s disease patients. PLoS One 2013;8:e59717. [Crossref] [PubMed]
- Schindlbeck KA, Eidelberg D. Network imaging biomarkers: insights and clinical applications in Parkinson’s disease. Lancet Neurol 2018;17:629-40. [Crossref] [PubMed]
- Eidelberg D, Moeller JR, Kazumata K, et al. Metabolic correlates of pallidal neuronal activity in Parkinson’s disease. Brain 1997;120:1315-24. [Crossref] [PubMed]
- Tzourio-Mazoyer N, Landeau B, Papathanassiou D, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002;15:273-89. [Crossref] [PubMed]
- Zhou H, Jiang J, Lu J, et al. Dual-model radiomic biomarkers predict development of mild cognitive impairment progression to Alzheimer’s disease. Front Neurosci 2019;12:1045. [Crossref] [PubMed]
- Haralick RM, Shanmugam K, Dinstein IH. Textural features for image classification. IEEE Trans Syst Man Cybern 1973.610-21. [Crossref]
- Galloway MM. Texture analysis using grey level run lengths. NASA STI/Recon Tech Rep N 1974:75.
- Chu A, Sehgal CM, Greenleaf JF. Use of gray value distribution of run lengths for texture analysis. Pattern Recognit Lett 1990;11:415-9. [Crossref]
- Dasarathy BV, Holder EB. Image characterizations based on joint gray level—run length distributions. Pattern Recognit Lett 1991;12:497-502. [Crossref]
- Thibault G, Fertil B, Navarro C, et al. Shape and texture indexes application to cell nuclei classification. Int J Pattern Recognit Artif Intell 2013;27:1357002. [Crossref]
- Amadasun M, King R. Textural features corresponding to textural properties. IEEE Trans Syst Man Cybern 1989;19:1264-74. [Crossref]
- Biau G, Scornet E. A random forest guided tour. Test 2016;25:197-227. [Crossref]
- Oshiro TM, Perez PS, Baranauskas JA. How many trees in a random forest? In: International workshop on machine learning and data mining in pattern recognition. Heidelberg: Springer, 2012:154-68.
- Orlhac F, Soussan M, Maisonobe JA, et al. Tumor texture analysis in 18F-FDG PET: relationships between texture parameters, histogram indices, standardized uptake values, metabolic volumes, and total lesion glycolysis. J Nucl Med 2014;55:414-22. [Crossref] [PubMed]
- Tatsumi M, Isohashi K, Matsunaga K, et al. Volumetric and texture analysis on FDG PET in evaluating and predicting treatment response and recurrence after chemotherapy in follicular lymphoma. Int J Clin Oncol 2019;24:1292-300. [Crossref] [PubMed]
- Becker AS, Ghafoor S, Marcon M, et al. MRI texture features may predict differentiation and nodal stage of cervical cancer: a pilot study. Acta Radiol Open 2017;6:2058460117729574. [Crossref] [PubMed]
- Klyuzhin IS, Blinder S, Mabrouk R, et al. Investigation of texture quantification parameters for neurological PET image analysis. In: 2015 IEEE nuclear science symposium and medical imaging conference (NSS/MIC). IEEE, 2015:1-5.
- Ben Bouallègue F, Vauchot F, Mariano-Goulart D, et al. Diagnostic and prognostic value of amyloid PET textural and shape features: comparison with classical semi-quantitative rating in 760 patients from the ADNI-2 database. Brain Imaging Behav 2019;13:111-25. [Crossref] [PubMed]
- Molina-García D, García-Vicente AM, Pérez-Beteta J, et al. Intratumoral heterogeneity in 18F-FDG PET/CT by textural analysis in breast cancer as a predictive and prognostic subrogate. Ann Nucl Med 2018;32:379-88. [Crossref] [PubMed]
- Acar E, Turgut B, Yiğit S, et al. Comparison of the volumetric and radiomics findings of 18F-FDG PET/CT images with immunohistochemical prognostic factors in local/locally advanced breast cancer. Nucl Med Commun 2019;40:764-72. [Crossref] [PubMed]
- Michoux N, Van den Broeck S, Lacoste L, et al. Texture analysis on MR images helps predicting non-response to NAC in breast cancer. BMC Cancer. 2015;15:574. [Crossref] [PubMed]
- Lewis MM, Galley S, Johnson S, et al. The role of the cerebellum in the pathophysiology of Parkinson’s disease. Can J Neurol Sci 2013;40:299-306. [Crossref] [PubMed]
- Teune LK, Renken RJ, de Jong BM, et al. Parkinson’s disease-related perfusion and glucose metabolic brain patterns identified with PCASL-MRI and FDG-PET imaging. Neuroimage Clin 2014;5:240-4. [Crossref] [PubMed]
- Teune LK, Renken RJ, Mudali D, et al. Validation of parkinsonian disease-related metabolic brain patterns. Mov Disord 2013;28:547-51. [Crossref] [PubMed]
- Niethammer M, Eidelberg D. Metabolic brain networks in translational neurology: concepts and applications. Ann Neurol 2012;72:635-47. [Crossref] [PubMed]
- Minen MT, Louis ED. Emergence of Parkinson’s disease in essential tremor: a study of the clinical correlates in 53 patients. Mov Disord 2008;23:1602-5. [Crossref] [PubMed]
- Lewis MM, Du G, Sen S, et al. Differential involvement of striato- and cerebello-thalamo-cortical pathways in tremor- and akinetic/rigid-predominant Parkinson’s disease. Neuroscience 2011;177:230-9. [Crossref] [PubMed]
- Sen S, Kawaguchi A, Truong Y, et al. Dynamic changes in cerebello-thalamo-cortical motor circuitry during progression of Parkinson’s disease. Neuroscience 2010;166:712-9. [Crossref] [PubMed]
- Yu H, Sternad D, Corcos DM, et al. Role of hyperactive cerebellum and motor cortex in Parkinson’s disease. Neuroimage 2007;35:222-33. [Crossref] [PubMed]
- Kwak Y, Peltier S, Bohnen NI, et al. Altered resting state cortico-striatal connectivity in mild to moderate stage Parkinson’s disease. Front Syst Neurosci 2010;4:143. [Crossref] [PubMed]
- Palmer SJ, Li J, Wang ZJ, et al. Joint amplitude and connectivity compensatory mechanisms in Parkinson’s disease. Neuroscience 2010;166:1110-8. [Crossref] [PubMed]


