Comprehensive and integrative analysis identifies COX7A1 as a critical methylation-driven gene in breast invasive carcinoma
Introduction
Breast cancer is the most widespread group of malignancies among females and the main reason for cancer-related death in the world (1). Breast invasive carcinoma is metastatic cancer and can commonly transfer to the lung, bone, liver, brain, and other organs, which primarily account for its incurability (2). In recent years, although molecular targeted therapy has improved treatment effect, the morbidity and mortality for patients with breast cancer are still increased year by year, and the risk increases annually (3). So, the underlying molecular biological mechanisms contribute to breast cancer carcinogenesis remains unclear.
It is now well recognized that one of the fundamental challenges in oncology is to detect the regulators of gene expression alterations during cancer development and progression. Promoter methylation is often hypothesized to be associated with reduced gene expression (4). Aberrant DNA methylation is a vital epigenetic modification as a hallmark of cancer and actively contributes to cancer progression by transcriptional silencing of critical tumor suppressor genes (5). The Cancer Genome Atlas (TCGA) is an extensive cancer genome project that archives and provides available information on mRNA expression, DNA methylation profiles, and so on (6), providing a tool to explore genes of interest or candidate biomarkers. The data of DNA methylation were obtained by using the Illumina Human Methylation 450 K BeadChip and downloaded from TCGA, which can detect nearly 450,000 methylation sites in the human genome with the single base resolution, covering 96% of CPG islands. Also, the beta value was applied to reflect the ratio of oligonucleotides that can match a given methylated sequence and the methylation rate in the sequence. Additionally, we utilized the R package MethylMix to provide a method that promotes the systematic analysis of methylation-driven genes in tumor studies (7). Based on the integrated analysis of methylation and gene expression profiles of TCGA data, we identified and validated a more comprehensive, reliable indicator in carcinogenesis of breast cancer and the potential role of DNA methylation. Moreover, we aimed to identify the expression patterns of methylation‐driven genes and the pathway involved in the development of breast cancer. Furthermore, our results also screened critical methylation-driven genes, potentially serving as the promising novel biomarkers for breast cancer diagnosis, prognosis, and treatment.
Methods
Data source and functional enrichment analysis
The expression and methylation data of breast invasive carcinoma (BRCA) patients were downloaded from the TCGA database (8). These data were normalized and analyzed to acquire the differentially expressed genes and aberrantly methylated genes by using the LIMMA package (9). Then, we selected the methylation-driven genes and obtained the correlation between gene expression level and gene methylation level by MethylMix algorithm.
DAVID was utilized to evaluate the common methylation-driven genes using GO and KEGG pathway enrichment analysis to analyze the biological processes, molecular functions, cellular components, and signaling pathways of these genes.
Cox proportional hazards model construction and model validation
The clinical information on BRCA patients was downloaded from the TCGA database. Then, univariate Cox proportional hazards regression analysis was used to identify candidate genes that were strongly involved with survival time. These genes with P value less than 0.05 were further employed in multivariate Cox proportional hazards regression analysis to screen hub genes. Next, we constructed the risk score model as follows: Risk Score (RS) = Σβi×expGenei (expGene: expression level of the gene, β: the regression coefficient). BRCA patients were assigned into high-risk and low-risk groups by using the median risk score as the cutoff point. Besides, Kaplan-Meier analysis was used to examine the proportional assumptions of the Cox proportional hazard model. By using R package survival ROC, receiver operating characteristic (ROC) was plotted and the area under the curve (AUC) was computed. Distribution of risk score, survival status, and gene expression levels were also plotted using the R software.
Tissue samples and cell culture
Fresh samples were obtained randomly from 26 BRCA patients and frozen at -80 °C until required for qPCR analysis, with informed consent. Our study was approved by the institutional ethics committee of the Affiliated Hospital of Nantong University.
Human breast cancer cell lines MDA-MB-468 and MDA-MB-231 were cultured in DMEM medium (Gibco, Carlsbad, CA, USA) with 10% FBS (Invitrogen, Carlsbad, CA, USA) and cultured in a humidified air atmosphere with 5% CO2 at 37 °C. Then, cells were seeded in a 6-well plate. After 24 h, the cell lines were treated with 5 and 10 µM of 5-Aza-2′-deoxycytidine (Aza) (Sigma, USA).
Quantitative reverse transcription PCR
By using TRIzol reagent (Invitrogen), the total RNA was extracted and collected from tissues and cell lines lysate. First-strand cDNA was synthesized from total RNA using a PrimeScript RT reagent kit (Takara, Japan). Then RT-PCR was applied with SYBR Green (Roche, Germany) on the Mastercycler Ep Realplex (Eppendorf 2S, Hamburg, Germany) according to standard protocols. Primers as follows: COX7A1-F: 5’-TGACATCCCGTTGTACCTGAA-3’; COX7A1-R: 5’-AAGGAGGCCCAGCCAAG-3’. The quantities of each mRNA were calculated using the 2−ΔΔCt method, and data were normalized using GAPDH as the loading control.
Methylation-specific PCR (MSP)
CpG island methylation status of the BRCA tissues was tested at the COX7A1 gene promoter regions by MSP. Genomic DNA was modified with bisulfite reagents. The modification converted unmethylated cytosine to thymine, whereas methylated cytosine stayed unchanged. After that bisulfite modified DNA was used to PCR amplification. If the CpG sites in the region are methylated, a methylated (M) band emerges. When the sites are unmethylated, the unmethylated (U) band is present. If the sites are partially methylated, both bands could appear. The MSP primers as follows: methylated: COX7A1-MSP-M-F: 5’-TGTAAAAATGTATTTTTTGGTATCGT-3’, COX7A1 MSP-M-R: 5’-AAATCCTACTCCTCGAATTCGTC-3’, and unmethylated: COX7A1-MSP-U-F: 5’-GTAAAAATGTATTTTTTGGTATTGT-3’, COX7A1-MSP-U-R: 5’-AAAAATCCTACTCCTCAAATTCATC-3’.
Statistical analysis
All survival analysis was used in R using the R package. GraphPad Prism 6 was used for data analysis. A t-test was performed for comparisons between two groups. P<0.05 was considered to be statistically significant.
Results
Identification of key methylation-driven genes in BRCA
Firstly, our study performed a model-based method named MethylMix that a beta mixture model to identify differential methylation status of genes. Then, we sought to get the DNA methylation data by TCGA data analysis of 436 BRCA samples and 54 normal samples. In addition, gene expression data were acquired from 1,053 cancer samples and 111 normal samples. According to the LIMMA software package, we extracted the gene expression and methylation expression data of BRCA from TCGA database. By using the MethylMix package, we performed the correlation analysis to screen methylation-driven genes by adopting the filter of P<0.05, |logFC| >0, and |Cor| >0.3. Finally, we acquired 218 methylation-driven genes in our study (Figure 1A).
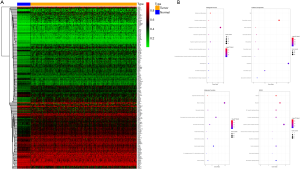
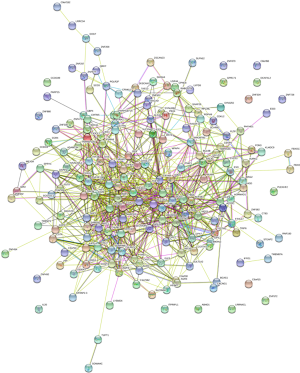
To further annotate the role of the above methylation-driven genes, we performed Gene Ontology (GO) and KEGG pathway enrichment analyzes. The GO results were significantly enriched in epithelial cell differentiation, xenobiotic catabolic process, regulation of transcription, DNA-templated, removal of superoxide radicals, extracellular exosome, and superoxide dismutase activity. Then the consequences of KEGG pathway analysis were significantly enriched in glioma, measles, and NF-kappa B signaling pathway (Figure 1B). Moreover, by using the STRING database, the PPI network among the methylation-driven genes was constructed with 190 nodes and 775 interactions (Figure S1).
Construction of methylation-driven genes-based prognostic signature
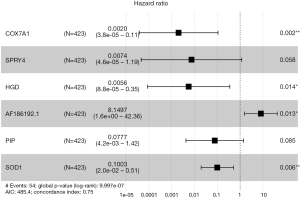
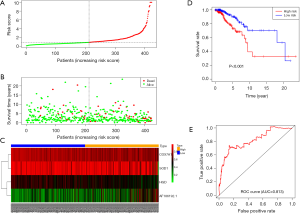
Univariate Cox regression analysis showed that 15 DEGs were related to the overall survival of patients (P<0.05). Stepwise multivariable Cox proportional hazards regression analyses were applied to identify a six-signature mRNA expression profile. Forest plot of hazard ratios revealed the prognostic values of these genes (Figure S2). Based on the hazards regression model, a final four genes were showed to be independently and significantly correlated with prognosis (P<0.05).
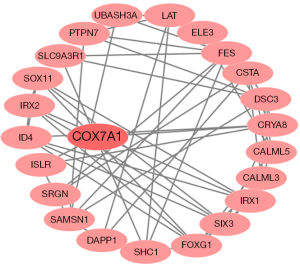
Patients were assigned to high-risk and low-risk groups. The risk scores of patients were distributed, and survival status was plotted for each patient on a dot plot (Figure 2A,B). The mortality in the high-risk group was much higher than that in the low-risk group. The heatmap showed the expression profiles of these four genes, and the expression profiles in the samples are ranked according to the risk score (Figure 2C). Furthermore, Kaplan-Meier risk curve revealed that patients in the high-risk group showed a dramatically shorter overall survival time than patients in the low-risk group (Figure 2D). ROC curve analysis for calculating survival in BRCA patients by the risk score was plotted and the AUC was 0.813, indicating good performance (Figure 2E). Given that Figure S3 presented the most significant module extracted from this PPI network by MCODE, we chose COX7A1 as a hub methylation-driven gene for further study.
Expression and prognostic assessment of COX7A1 in BRCA
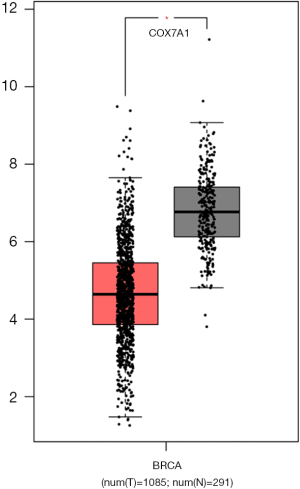
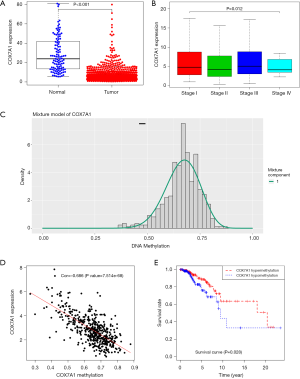
A total of 1164 BRCA samples with COX7A1 expression data across all patient characteristics were analyzed from TCGA. We verified the expression level of COX7A1 using LIMMA software in TCGA data and found that down-regulation of COX7A1 expression was in tumor tissues compared with normal tissues (Figure 3A, P<0.001). Also, decreased expression of COX7A1 correlated obviously with the tumor clinical stage (Figure 3B, P=0.012), which suggested that BRCAs with low COX7A1 expression are prone to progress to a more advanced stage than those with high COX7A1 expression. To further confirm the above results, we tested the COX7A1 expression by using the GEPIA database (Figure S4). To validate the function of methylation in regulating the expression of COX7A1, we performed the MethylMix model to exhibit the low methylation state corresponds to the normal methylation and the high methylation state matches hypermethylation of COX7A1 (Figure 3C). Moreover, the inverse correlation between DNA methylation and matched COX7A1 gene expression (Figure 3D, correlation coefficient: −0.686). To furthermore evaluate the prognostic value of methylation-driven gene, Kaplan-Meier survival curve analysis of patients in the hypermethylation and hypomethylation groups indicated that the overall survival rate was higher in the hypermethylation group (Figure 3E, P=0.028), which showed that COX7A1 was independent prognostic indicators for BRCA.
COX7A1 expression level is regulated by DNA methylation
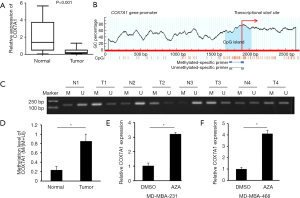
We further detected the COX7A1 expression level in BRCA tissues and found that the COX7A1 expression level in tumor tissues was significantly decreased compared with corresponding normal tissues by qPCR analysis (P<0.05; Figure 4A). To investigate whether DNA methylation was responsible for COX7A1 expression, we searched the status of CpG islands in the COX7A1 promoter using MethPrimer software and found significant CpG islands (Figure 4B). We further analyzed the CpG island methylation status in the COX7A1 promoter in tumor tissues and matched normal tissues from 18 BRCA patients. The results of MSP showed that the methylation level in the promoter of the COX7A1 in BRCA tissues was higher than that in paired normal tissues (P<0.05, Figure 4C,D).
To further verify whether low expression of COX7A1 in BRCA is attributed to DNA hypermethylation, we treated MDA-MB-468 and MDA-MB-231 cells with the DNA methylation inhibitor 5-Aza-dC (10), and then evaluated COX7A1 expression. The results of qPCR analysis exhibited that there was an obvious increase in COX7A1 levels in both MDA-MB-468 and MDA-MB-231 cells after treated with 5-Aza-dC (Figure 4E,F), which suggested that COX7A1 expression in BRCA cells might be regulated by DNA methylation.
The landscape of tumor-infiltrating immune cells in BRCA with low and high expression of COX7A1
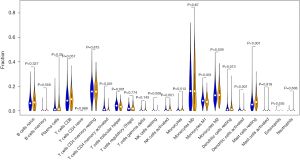
In this study, based on the TCGA BRCA dataset, we further estimated the abundance of 24 immune cell subsets in high and low expression of COX7A1 by using the CIBERSORT algorithm (11). The tumor infiltrating immune cells including major types associated with T cells follicular helper, T cells CD4 memory activated, NK cells resting, NK cells activated, Macrophages M1, Dendritic cells activated, Dendritic cells resting, and Mast cells resting, showed significantly differential enrichments in low- and high-expression of COX7A1 (Figure 5, P<0.05). These findings suggested that expression levels of COX7A1 are associated with immune activity tumor infiltrating immune cells.
Discussion
Previous reports indicated that aberrant epigenetic changes played a crucial roles in breast cancer progression (12). Epigenetic change such as DNA methylation is a vital mechanism that regulates gene transcription, and its effects in breast cancer development have been studied widely (13). In addition, methylated genes may also be attractive treatment targets in breast cancer using therapies in trials in other tumour types (14). So, new biomarker discovery is crucial for diagnostic innovation and personalized medicine in breast cancer. Microarray and high-throughput sequencing technologies derived from TCGA provide effective tools for analyzing key genetic or epigenetic changes in the genesis and development of cancer (15).
In the current study, a previously published database TCGA was used to identify significant methylation-driven genes. A total of 218 genes were revealed in BRCA. Then, we further analyzed the biological process, molecular function, and cellular component of these methylation-driven genes using GO and KEGG pathway enrichment analysis in order to explore their role in the genesis and development of BRCA. Interestingly, the GO results were predominantly enriched in epithelial cell differentiation, xenobiotic catabolic process, regulation of transcription, DNA-templated, removal of superoxide radicals, extracellular exosome, and superoxide dismutase activity. And the results of KEGG pathway analysis were also revealed to be enriched in glioma, measles, and NF-kappa B signaling pathway. A risk scoring system of four signature expression profiles was established by using univariate and stepwise multivariate Cox regression analyses, including COX7A1, SOD1, HGD, and AF186192.1. In this study, COX7A1 was chosen as a candidate because that down-regulated expression of COX7A1 was only significantly related to BRCA stage. Stage I and II BRCA patients showed higher COX7A1 expression, and stage IV BRCA patients showed lower COX7A1 expression. Therefore, the expression patterns and methylation status of COX7A1 remain to be confirmed and validated in future studies.
COX7A1 gene, encoding cytochrome c oxidase subunit VII polypeptide 1, is located on chromosome 19q13.12 within a CpG dense region (16), which show differential DNA methylation (17). Promoter CpG island methylation is one of the major driver events that play important roles during tumor progression (18). Previous studies have reported that COX7A1 is most abundantly expressed in heart and skeletal muscle (19), and is markedly reduced in diabetic muscle (20). In recent years, COX7A1 expression was reported to be down-regulated in lung cancer tissues and COX7A1 overexpression in A549 cells resulted in the suppression of cell proliferation and enhance in cell death via apoptosis (21). In this study, our results found that down-regulated expression of COX7A1 in BRCA tissue samples, and significantly associated with clinical stage. MethylMix was used to identify DNA methylation driven genes by modeling DNA methylation data in tumor and normal (7). Based on MethylMix model, we further identified COX7A1 as a methylation-driven gene. And we found that the methylation level of COX7A1 negatively correlated with its expression level, and closely related to the patients’ survival rate with BRCA. Moreover, our validation exhibited that the expression patterns and methylation status of COX7A1 by western blot and MSP. As is well-known, DNA methylation is predominant epigenetic mechanisms that downregulate tumor suppressor genes in cancers (22). Thus, we treated breast cancer cell lines with 5-Aza-dC and found that the COX7A1 expression level was markedly elevated in MDA-MB-468 and MDA-MB-231 cell lines after 5-Aza-dC treatment, which showed that abnormal methylation contributed to the inactivation of COX7A1 in BRCA cell lines. In addition, based on CIBERSORT, we also performed a comprehensive analysis of the COX7A1 expression on the tumor-infiltrating immune cells in BRCA.
Conclusions
In summary, through integrated bioinformatics analysis and in vitro experiment, our results suggested that the pathogenesis of BRCA may result from epigenetically regulated expression levels of COX7A1. Together, these findings collectively demonstrated the critical role of COX7A1 as a potential therapeutic target for BRCA, and also provided new insights into the mechanism of breast cancer.
Acknowledgments
Funding: This study was supported by the Social Development Foundation of Nantong City (MS12017007-1) and the National Natural Science Youth Foundation of China (No.81702608).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Our study was approved by the ethics committee of the Affiliated Hospital of Nantong University (No. 2013-067).
References
- Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Sun YS, Zhao Z, Yang ZN, et al. Risk Factors and Preventions of Breast Cancer. Int J Biol Sci 2017;13:1387-97. [Crossref] [PubMed]
- Bcj M, Boyce AM, Jvm GB, et al. Increased Risk of Breast Cancer at a Young Age in Women with Fibrous Dysplasia. J Bone Miner Res 2018;33:84-90. [Crossref] [PubMed]
- Baylin SB, Esteller M, Rountree MR, et al. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet 2001;10:687-92. [Crossref] [PubMed]
- Gerda E, Gangning L, Ana A, et al. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004;429:457-63. [Crossref] [PubMed]
- Lee JS. Exploring cancer genomic data from The Cancer Genome Atlas Project. BMB Rep 2016;49:607-11. [Crossref] [PubMed]
- Gevaert O. MethylMix: an R package for identifying DNA methylation-driven genes. Bioinformatics 2015;31:1839-41. [Crossref] [PubMed]
- Hutter CM, Zenklusen JC. The Cancer Genome Atlas: Creating Lasting Value beyond Its Data. Cell 2018;173:283-5. [Crossref] [PubMed]
- Diboun I, Wernisch L, Orengo CA, et al. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics 2006;7:252. [Crossref] [PubMed]
- Bender CM, Zingg J, Jones PA. DNA methylation as a target for drug design. Pharm Res 1998;15:175-87. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Sato F, Saji S, Toi M. Genomic tumor evolution of breast cancer. Breast Cancer 2016;23:4-11. [Crossref] [PubMed]
- Johnson KC, Koestler DC, Fleischer T, et al. DNA methylation in ductal carcinoma in situ related with future development of invasive breast cancer. Clin Epigenetics 2015;7:75. [Crossref] [PubMed]
- Issa JP. DNA Methylation as a Therapeutic Target in Cancer. Clin Cancer Res 2007;13:1634-7. [Crossref] [PubMed]
- Stoll D, Akbani R. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059-74. [PubMed]
- Wolz W, Kress W, Mueller CR. Genomic Sequence and Organization of the Human Gene for Cytochrome c Oxidase Subunit (COX7A1) VIIa-M. Genomics 1997;45:438-42. [Crossref] [PubMed]
- Chalaya TV, Akopov SB, Nikolaev LG, et al. Tissue specificity of methylation of cytosines in regulatory regions of four genes located in the locus FXYD5-COX7A1 of human chromosome 19: Correlation with their expression level. Biochemistry (Mosc) 2006;71:294-9. [Crossref] [PubMed]
- Costello JF, Fruhwald MC, Smiraglia DJ, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet 2000;24:132-8. [Crossref] [PubMed]
- Jaradat SA, Ko MSH, Grossman LI. Tissue-Specific Expression and Mapping of the Cox7ah Gene in Mouse. Genomics 1998;49:363-70. [Crossref] [PubMed]
- Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1 alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267-73. [Crossref] [PubMed]
- Mishra N, Timilsina U, Ghimire D, et al. Downregulation of cytochrome c oxidase subunit 7A1 expression is important in enhancing cell proliferation in adenocarcinoma cells. Biochem Biophys Res Commun 2017;482:713-9. [Crossref] [PubMed]
- Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet 2007;8:286-98. [Crossref] [PubMed]