
The in vitro activity of polymyxin B and tigecycline alone and combination with other antibiotics against carbapenem-resistant Enterobacter cloacae complex isolates, including high-risk clones
Introduction
The World Health Organization (WHO) has listed carbapenem-resistant enterobacteria (CRE) as one type of bacteria that is in urgent need of new antibiotics (Abs) development. Most in vitro CRE data concerns Klebsiella pneumoniae with Enterobacter cloacae complex (ECC) data being poorly understood (1). With the emergence of the carbapenem-resistant Enterobacter cloacae complex (CREC), combination antimicrobial therapies are more often being considered. Polymyxin B (PMB) Abs have been used since the late 1950s, but can cause ototoxicity and neurotoxicity; less toxic aminoglycosides in the 1970s gradually replaced PMB Abs. However, due to the lack of new Ab development, PMBs continued to be used in clinical practice, even with the pharmacokinetic/pharmacodynamic limitations and increased risk of severe renal insufficiency. It was found that PMBs might promote membrane transport of other Abs, thereby enhancing bactericidal or bacteriostatic activities (2,3). The broad-spectrum antimicrobial agent, tigecycline (TGC), was also active against most Gram-negative isolates in vitro, including ESBL producers. TGC is indicated for the treatment of complicated skin and soft tissue infections (4). PMB or TGC has been used in Ab combination therapy studies (2,5,6). However, past in vitro studies has ignored the importance of using clinically achievable concentrations to determine antibacterial effects. In our study, we used the checkerboard method and time-kill analysis (TKAs) to evaluate the effect of PMB or TGC combined with meropenem (MEM), amikacin (AMK), and levofloxacin (LEV) against seven CREC isolates. We also studied the effects of TGC combined with PMB. The purpose of this study was to provide more guidance for the medical treatment of CREC infection.
Methods
Bacterial isolates
Seven non-repeated clinical CREC strains were collected from the First Affiliated Hospital of Jiamusi University between September 2016 and December 2018, and all isolates were stored frozen until further use. Species identifications and minimum inhibitory concentrations (MICs) of the Abs evaluated were analyzed using the Vitek2 compact system (BioMerieux, France). MICs of MEM, PMB, TGC, AMK, and LEV were further determined by broth microdilution according to the guidelines of the 2016 Clinical and Laboratory Standards Institute (CLSI 2016).
Antibiotics
Antimicrobial agents included PMB (Sigma Chemical, St. Louis, MO, USA), AMK (Tianjin Chemical Co., Ltd., Tianjin, China), LEV (CISEN Chemical Industries, Ltd., Shandong, China), MEM (Haibin Chemical Industry Co. Ltd., Shenzhen, China), and TGC (Pfizer Co. Ltd., Hangzhou, China). A PMB concentration of 2 µg/mL is considered a sensitive breakpoint (7).
PCR studies
The resistance genes were detected using the polymerase chain reaction (PCR), including MBL (blaNDM-1, blaIMP-4, blaVIM) and others β-lactamases (blaTEM, blaSHV, blaCTX-15, blaCTX-9, blaKPC, blaOXA-48, blaOXA-1,blaOXA-23,blaOXA-24, blaOXA-51, and blaOXA-58). AmpC enzyme genes (blaACC, blaDHA, blaCMY), quinolone resistance determining regions (QRDRs), gyrA and parC, and plasmid-mediated quinolone resistance (PMQR) determinants [qnrA, qnrB, qnrS, aac(6’)-Ib, and qepA] (8,9) were also detected. The colistin resistance gene mcr-1 primer sequences and conditions were previously reported by Liu et al. (10).
Multilocus sequence typing (MLST)
MLST for seven housekeeping genes, including dnaA, fusA, gyrB, leuS, pyrG, rplB, and rpoB, were performed according to a previously described method (https://pubmlst.org/ecloacae/).
Synergy evaluations using the checkerboard assay
The activities of PMB + TGC, and PMB or TGC combined with AMK, LEV, and MEM were measured against seven bacterial strains derived from FICI values using the microdilution checkerboard method. Five Ab concentrations were prepared, 1/8× MIC, 1/4× MIC, 1/8× MIC, 1× MIC, and 2× MIC. Along with the horizontal and vertical wells, 50 µL of each Ab was added into each well, and then a 5×105 CFU/mL bacterial suspension was inoculated into each well. The negative control wells contained only MH broth (MHB). The results were interpreted as follows: a FICI ained only MH bro, 0.5< FICI ≤4, indifferent, and FICI >4, antagonistic (11). E. Cloacae (National Center for Medical Culture Collections, CMCC-45301) were used as quality control strains.
The minimum bactericidal concentration (MBC) method of detection
An inoculum that contained 5×105 CFU/mL bacterial suspensions in the exponential growth phase was used for these experiments. Microtiter plates were inoculated with the bacterial suspension and diluted antimicrobials. After incubating the plates at 37 °C for 24 h, suspensions corresponding to 1×, 2×, 4×, and 8× the MICs of the compound were spread onto MH agar plates and incubated overnight at 37 °C. Plates that had a colony counts of less than 10 were considered to be the MBC (12).
Time-kill assays (TKA)
TKA were conducted for each strain according to previously described methods. The Ab concentrations used for PMB were 0.25 and 1 µg/mL. The Ab concentrations used for TGC were 0.25 and 1 µg/mL. The concentrations for MEM with PMB or TGC were 4 and 16 µg/mL. For AMK with PMB or TGC, the concentrations were 8 and 16 µg/mL. The concentrations of LEV with PMB or TGC were 6.4 µg/mL. All concentrations were determined according to clinically achievable concentrations (7,13). A 5×105 CFU/mL inoculum of the tested organism with into 10 mL of the corresponding broth (containing the Abs alone or in combination). The samples were obtained aseptically at 0, 2, 4, 8, and 24 hours. Regrowth was defined as an increase in colony counts from the previous time point. Synergistic effects were determined by a decrease of ≥2 log10 CFU/mL when compared with the combined Abs, with the most active single antimicrobial alone at that time point, while an increase of >2 log10 was considered antagonistic.
Results
Antibiotic susceptibility and level of resistance
Susceptibilities for seven strains are listed in Table 1. In summary, all isolates were susceptible to PMB (MICs were 2–4 µg/L), MEM (MICs were 8–256 µg/L); AMK (MICs were 0.5–20,480 µg/L), TGC (MICs were 0.25–1 µg/L), and LEV (MICs were 0.0625–32 µg/L). Four strains that were resistant to PMB (MIC =4 µg/L), but a mcr-1 gene was not found.
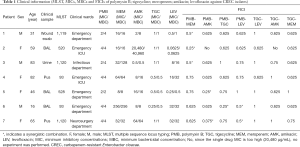
Full table
Epidemiological links and drug resistance gene of CREC
Seven isolates were clustered into five sequence types (STs), including ST520, ST528, ST1119, ST1120, and ST93. Among them, ST1119 and ST1120 are newly discovered STs (Table 1). The clustering of CREC isolates was also apparent in the analysis of the antimicrobial resistance gene contents, as shown in Figure 1. Similar ST types had a similar drug resistance phenotype. The isolates were classified according to the content of resistance determinants using a binary distance matrix and the UPGMA clustering method. In brief, except for the CREC6 isolate, all strains carried the metal β-lactamase gene. Interestingly, the CREC3 strain co-produced the blaNDM-1 and blaIMP-4 genes. The blaOXA-1 carbapenemase class D family gene was detected in four isolates. Among the AmpC-producing enzymes, blaACC and blaDHA were found. Focusing on PMQR harbored by these isolates, we showed that the qnr and aac(6’)-Ib-cr genes were detected. Two qnr family genes were also found (qnrB and qnrS), no qnrA or qepA genes were detected, and QRDR DNA sequences of the gyrA and parC genes were identified. Mutations were seen in three sites of the CREC4 strain and the two sites of the CREC6 strain. The MICs of the two isolates were 16 and 32 µg/mL, respectively. In addition, mutations in the one site of the CREC3 and CREC7 strains were found, and the MIC of the two isolates were 8 µg/mL. Substitutions in the QRDR were detected in LEV resistant strains, as shown in Figure 1.
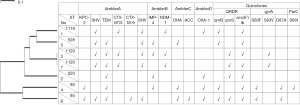
Checkerboard synergy analysis
The results of the checkerboard combination studies are shown in Table 1. The combination of PMB and MEM resulted in FICI values of 0.5, 0.25, and 0.5 against CREC1, CREC2, and CREC3, respectively. The combination of PMB and AMK resulted in FICI values of 0.25 and 0.375 against CREC5 and CREC7, respectively. PMB and LEV displayed FICI values of 0.5 against CREC6 and CREC7, and PMB + TGC resulted in FICI values of 0.25 against CREC6, which indicated synergistic interactions. The results of the checkerboard combination studies based on TGC showed no synergistic interactions. It is worth noting that antagonism was not found with any combination. No MBC values exceeded 4× MIC, as shown in Table 1.
Time-kill assays results
All monotherapies were evaluated against the seven isolates using the TKAs. Specifically, MEM used alone at 4, and 16 µg/mL concentrations were bacteriostatic, resulting in regrowth by 8 hours for all isolates. PMB used alone at 0.25, and 1 µg/mL was rapidly bactericidal for two isolates by 2 hours (the CREC1 and CREC5 strains). Other strains exhibited bacteriostatic actions, and regrowth was observed by 8 hours except for the CREC1 strain. TGC used alone at 0.25, and 1 µg/mL was bacteriostatic for all isolates by 2 hours, but regrowth was observed by 8 hours. AMK used alone at 16 µg/mL was bactericidal by 8 hours for the CREC1, CREC3, and CREC6 isolates; for these three strains, bactericidal activity was maintained for 24 hours. LEV used alone at 6.8 µg/mL was bactericidal by 24 hours for the CREC1 and CREC2 isolates; log change = logCFU24 – logCFU0, as shown in Figure 2.

Because the monotherapies were effectively bactericidal against the CREC1, CREC2, CREC3, and CREC6 isolates, we selected three clinical isolates (CREC4, CREC5, and CREC7) that the monotherapies were not bactericidal against and evaluated the in vitro activity of PMB + TGC and PMB or TGC in combinations with MEM, AMK, and LEV.
In the TKA, synergy was not observed for all of the combinations. In general, PMB-based combination had a better antibacterial effect against the three isolates compared with the TGC-based combinations.
For CREC4 isolate, PMB+AMK combination had a better antibacterial effect with the TGC+AMK combinations. TGC + MEM had a bacteriostatic and synergistic effect against CREC4 and increasing MEM concentrations had more pronounced synergistic effects. However, for the PMB + MEM combination, synergistic interactions were not observed. TGC + LEV appeared to be indifferent against the CREC4 isolates (Figure 3A,B,C,D).
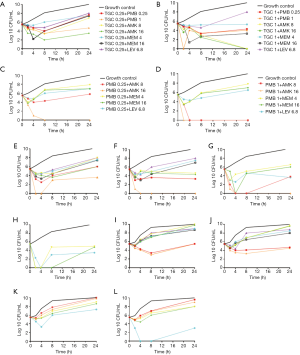
For CREC5 isolate, PMB + AMK combination also had a better antibacterial effect with the TGC + AMK combinations. The TGC + MEM and PMB + MEM synergistic interactions were not observed. TGC + LEV and PMB + LEV appeared to be indifferent against the CREC5 isolates (Figure 3E,F,G,H).
For CREC7 isolate, the PMB + AMK and TGC + AMK combination had no bactericidal effects, for the PMB + MEM and TGC + MEM combination, synergistic interactions were not observed. High PMB + LEV concentrations have bacteriostatic and synergistic effects against the CREC7 isolate (Figure 3I,J,K,L).
We also combined PMB + TGC, which showed synergistic effects against all three strains; however, the bactericidal effects were not obvious, as shown in Table 2 and Figure 3.
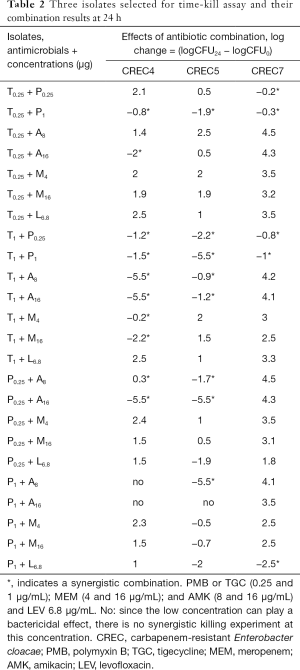
Full table
Discussion
The presence of CRECs is due to the loss of porin, overproduction of a chromosomally encoded cephalosporinase, AmpC, activation of the efflux system, and the presence of important carbapenem hydrolases, such as metallo-beta-lactamase and blaKPC (14). In China, the first reported CREC was a KPC-producing strain isolated from Shanghai in 2010 (15). Later, the CREC blaNDM, blaIMP, and blaVIM genes were reported from different geographical regions (16). A previous CREC study showed that KPC-producing ECC exhibited lower MEM MICs of 2–8 µg/mL (7). In our study, KPC-producing CREC isolates exhibited higher MEM MICs (64–256 µg/mL) than IMP- and NDM-producing isolates (16–32 µg/mL). Fluoroquinolone (FQ) resistance is caused by mutations in DNA gyrase and topoisomerase IV chromosomal genes, and DNA gyrase is the primary target enzyme where substitutions occur to resist FQs. Our research results show that alterations in both GyrA and ParC often confer high-level resistance, consistent with conclusions by Hooper et al. (17).
Peirano et al. reported that ST93 had a global distribution and was associated with different carbapenemases (blaIMP-8, blaIMP-14, blaVIM-1, blaNDM-1, blaKPC-2, and blaOXA-48) (18). Of the seven CREC isolates studied, two carried the blaKPC-2 carbapenemase and belonged to ST93. Interestingly, although the two isolates have similar resistant phenotypes, the effects of antibiotic combination therapy vary widely. In addition, results from another study confirmed that antibacterial activity is strain-specific, consistent with our findings. In a recent Brazilian study, a high mortality rate associated with KPC-producing Enterobacter cloacae was found in 24 patients, where the mortality rate reached 41.8% (19). Therefore, for the ST93 KPC-2-producing high-risk clones, infection control measures should be performed, and stricter Ab policies should be required to control the spread of ST93.
The cost to treat CRE is higher than with other chronic diseases. A study evaluating the health and economic burden of CRE in the United States showed that, depending on the infection type, the cost to the family, hospital, and society for a single CRE infection had created a substantial economic burden (20). And, while the number of studies is increasing, exploration of effective combinations against CRE are few. Combination therapies include the follow advantages: (I) synergistic effects have been demonstrated in in vitro studies that showed better clinical outcomes, and helped to guide clinical decisions, (II) drug resistance could rapidly develop if a sole antimicrobial, such as PMB and TGC, is given, (III) combination drug therapy could reduce the dose of each Ab, reducing side effects and decreasing patient costs.
Combining antimicrobials with different mechanisms of action could provide better pharmacokinetic effects. In contrast to most previous studies, which employed colistin in combination with other Abs against CREC, we chose PMB as the basis for the antimicrobial combinations because of its lower nephrotoxicity and higher tissue concentrations compared with colistin (2). Randomized trials have indicated that using TGC alone could confer an increased mortality risk (21), which further illustrates that TGC is best suited for combination therapy.
The lack of a gold standard has brought significant challenges in being able to detect synergistic effects on the clinical microbiology laboratory using in vitro drug susceptibility. TKA is time-consuming and technically challenging, but articles about TKA are often considered more relevant for clinical situations (22). The checkerboard method measures only the inhibitory activity. E-test synergy methods are more rapid and easier to perform, but there are more affected by other factors. In our research, the checkerboard and TKA methods are different. The results of the TGC-based checkerboard method were indifference, but with the TKS method, the TGC-based checkerboard method showed synergy with AMK and MEM. At present, based on the limited data available, it is not possible to decide which in vitro test method is the most relevant for clinical outcomes, so further research is urgently needed. It is worth noting that with the TKA, the superiority of PMB + MEM in combination with CREC was not found, but the synergistic effect against the CREC4 isolate was found with TGC + MEM. However, in a recent clinical study, TGC + MEM produced an antagonistic effect in the treatment of a KPC-2-producing Klebsiella pneumonia infection, and eventually, the patient was cured with an increased dose of TGC (23). This result is inconsistent with our study. In our study, we found that TGC + MEM had a synergistic effect on the CREC4 isolate producing KPC-2, and no antagonism was found. The likely reason is that the combined effect differs among different strains. In addition, further clinical studies should be performed for the combination of TGC + MEM.
Limited studies have explored the activity of PMB or TGC + AMK against CREC. Cai et al. reported that PMB + AMK was synergistic against PMB-sensitive Enterobacter cloacae in 2/4 cases; however, no synergistic effect was evident against the PMB-resistant strains (2). Therefore, we evaluated KPC-producing PMB-resistant CREC isolates and found that PMB + AMK had bactericidal and synergistic activity against AMK-susceptible CREC isolates that produce KPC and/or IMP-4. Compared with PMB, TGC requires high concentrations to act on AMK-sensitive strains (CREC4). Therefore, the combination of PMB + AMK is promising for the treatment of AMK-sensitive CREC.
Cai and Betts et al. found that PMB + TGC was promising in the treatment of CREC (2,24). In contrast, PMB and TGC (at 0.25 or 1 µg/mL) were used in combination with other Abs, and we did not find sustained bactericidal activity against all three CREC isolates within 24 hours except for PMB-sensitive CREC5. Therefore, TGC + PMB should be used with caution, against the PMB-resistant CREC. The CREC7 strain was resistant to both PMB, AMK, and LEV, so no bactericidal effect was observed for all of our experimental combinations. Only high PMB concentrations combined with LEV showed synergistic and bacteriostatic effects.
The antimicrobial concentrations used in the tests are important when considering the antimicrobials combinations assays used. In this study, we used clinically achievable concentrations to determine antibacterial effects, making the obtained results more useful to clinical medicine. Our study has some limitations. First, the number of isolates was small, and those in Jiamusi, China, only represented the hospitals. Second, further clinical studies determining the relevance of these data are warranted.
Conclusions
In this study, we showed that the ST93 of the KPC-2 producing high-risk clones are dangerous, and research should be intensified to combat these bacteria with stricter Ab policy requirements to control the spread of CREC. In addition, we found that the combination of PMB + AMK is promising for the treatment of AMK-sensitive CREC. Future studies should investigate these drug combinations in animal models, and studies of the pharmacokinetic parameters of these drug combinations should be performed.
Acknowledgments
Funding: This work was supported by the Excellent Team of Young Teachers Foundation of Heilongjiang Province (2018-KYYWF-0916) and Heilongjiang Provincial Health and Family Planning Commission on Scientific Research Project (2018-290) and Research Project of Yongchuan Hospital Affiliated to Chongqing Medical University (YJYJ201902).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The Ethics Committee of Jiamusi University Clinical Medical College approved the study protocol for research. The committee’s reference number is 20180326. Individual informed consent was waived by the ethics committee listed above because this study used currently existing samples collected during routine medical care and did not cause any additional risks to the patients.
References
- Cebrero-Cangueiro T, Alvarez-Marin R, Labrador-Herrera G, et al. In vitro Activity of Pentamidine Alone and in Combination With Aminoglycosides, Tigecycline, Rifampicin, and Doripenem Against Clinical Strains of Carbapenemase-Producing and/or Colistin-Resistant Enterobacteriaceae. Front Cell Infect Microbiol 2018;8:363. [Crossref] [PubMed]
- Cai Y, Lim TP, Teo J, et al. In Vitro Activity of Polymyxin B in Combination with Various Antibiotics against Extensively Drug-Resistant Enterobacter cloacae with Decreased Susceptibility to Polymyxin B. Antimicrob Agents Chemother 2016;60:5238-46. [Crossref] [PubMed]
- Poulakou G, Lagou S, Karageorgopoulos DE, et al. New treatments of multidrug-resistant Gram-negative ventilator-associated pneumonia. Ann Transl Med 2018;6:423. [Crossref] [PubMed]
- Decousser JW, Woerther PL, Soussy CJ, et al. The tigecycline evaluation and surveillance trial; assessment of the activity of tigecycline and other selected antibiotics against gram-positive and gram-negative pathogens from France collected between 2004 and 2016. Antimicrob Resist Infect Control 2018;7:68. [Crossref] [PubMed]
- Barth N, Ribeiro VB, Zavascki AP. In vitro activity of polymyxin B plus imipenem, meropenem, or tigecycline against KPC-2-producing Enterobacteriaceae with high MICs for these antimicrobials. Antimicrob Agents Chemother 2015;59:3596-7. [Crossref] [PubMed]
- Ozbek B, Mataraci-Kara E, Er S, et al. In vitro activities of colistin, tigecycline and tobramycin, alone or in combination, against carbapenem-resistant Enterobacteriaceae strains. J Glob Antimicrob Resist 2015;3:278-82. [Crossref] [PubMed]
- Kulengowski B, Rutter WC, Campion JJ, et al. Effect of increasing meropenem MIC on the killing activity of meropenem in combination with amikacin or polymyxin B against MBL- and KPC-producing Enterobacter cloacae. Diagn Microbiol Infect Dis 2018;92:262-6. [Crossref] [PubMed]
- Gong X, Zhang J, Su S, et al. Molecular characterization and epidemiology of carbapenem non-susceptible Enterobacteriaceae isolated from the Eastern region of Heilongjiang Province, China. BMC Infect Dis 2018;18:417. [Crossref] [PubMed]
- Rodriguez-Martinez JM, Velasco C, Pascual A, et al. Correlation of quinolone resistance levels and differences in basal and quinolone-induced expression from three qnrA-containing plasmids. Clin Microbiol Infect 2006;12:440-5. [Crossref] [PubMed]
- Liu Y-Y, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 2016;16:161-8. [Crossref] [PubMed]
- Odds FC. Synergy, antagonism, and what the chequerboard puts between them. J Antimicrob Chemother 2003;52:1. [Crossref] [PubMed]
- French GL. Bactericidal agents in the treatment of MRSA infections--the potential role of daptomycin. J Antimicrob Chemother 2006;58:1107-17. [Crossref] [PubMed]
- Geerdes-Fenge HF, Wiedersich A, Wagner S, et al. Levofloxacin pharmacokinetics and serum bactericidal activities against five enterobacterial species. Antimicrob Agents Chemother 2000;44:3478-80. [Crossref] [PubMed]
- Kalil AC, Metersky MM. Are there more differences or similarities between the hospital-acquired pneumonia guidelines? Ann Transl Med 2018;6:429. [Crossref] [PubMed]
- Wu Q, Liu Q, Han L, et al. Plasmid-mediated carbapenem-hydrolyzing enzyme KPC-2 and ArmA 16S rRNA methylase conferring high-level aminoglycoside resistance in carbapenem-resistant Enterobacter cloacae in China. Diagn Microbiol Infect Dis 2010;66:326-8. [Crossref] [PubMed]
- Jin C, Zhang J, Wang Q, et al. Molecular Characterization of Carbapenem-Resistant Enterobacter cloacae in 11 Chinese Cities. Front Microbiol 2018;9:1597. [Crossref] [PubMed]
- Hooper DC. Mechanisms of fluoroquinolone resistance. Drug Resist Updat 1999;2:38-55. [Crossref] [PubMed]
- Peirano G, Matsumura Y, Adams MD, et al. Genomic Epidemiology of Global Carbapenemase-Producing Enterobacter spp., 2008-2014. Emerg Infect Dis 2018;24:1010-9. [Crossref] [PubMed]
- da Silva KE, Varella TR, Bet G, et al. High mortality rate associated with KPC-producing Enterobacter cloacae in a Brazilian hospital. Am J Infect Control 2018;46:108-10. [Crossref] [PubMed]
- Bartsch SM, McKinnell JA, Mueller LE, et al. Potential economic burden of carbapenem-resistant Enterobacteriaceae (CRE) in the United States. Clin Microbiol Infect 2017;23:48.e9-48.e16. [Crossref] [PubMed]
- Tasina E, Haidich AB, Kokkali S, et al. Efficacy and safety of tigecycline for the treatment of infectious diseases: a meta-analysis. Lancet Infect Dis 2011;11:834-44. [Crossref] [PubMed]
- Eliopoulos GM, Eliopoulos CT. Antibiotic combinations: should they be tested? Clin Microbiol Rev 1988;1:139-56. [Crossref] [PubMed]
- Bi S, Yao X, Huang C, et al. Antagonistic effect between tigecycline and meropenem: successful management of KPC-producing Klebsiella pneumoniae infection. Infection 2019;47:497-500. [Crossref] [PubMed]
- Betts JW, Phee LM, Hornsey M, et al. In vitro and in vivo activities of tigecycline-colistin combination therapies against carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother 2014;58:3541-6. [Crossref] [PubMed]


