
Emerging biomarkers in autism spectrum disorder: a systematic review
Introduction
Autism spectrum disorder (ASD) is perhaps one of the most important medical disorders of our era because of the number of people it affects. The prevalence of ASD has increased significantly over the past three decades and now is estimated to affect 2% (1) or more (2) of children in the United States (US). Even more significant is the fact that ASD does not occur in isolation. Children with ASD require significant support from the educational, medical and social systems that results in a significant economic burden (3) which is estimated to cost the US approximately $268 billion in 2015 (4). In addition, the disability of a child creates a spillover effect, decreasing the quality of life for the entire family (5-7).
One of the major limitations when identifying, diagnosing, treating and understanding ASD is the fact that ASD is exclusively defined based on the observation of behaviors by trained or untrained individuals. The complications resulting from this approach are discussed in more detail below, but the fact remains that there is no proven biological measurement (e.g., blood test or radiological scan) which can identify pathophysiological processes that can aid in the diagnosis or treatment of ASD at this time. However, many promising areas of research have uncovered abnormal biological processes that are associated with ASD. Biomarkers developed to measure these biological abnormalities accurately could be important in the diagnosis and management of ASD.
Biomarkers are objective measures of biological or pathophysiological processes, or pharmacologic responses to therapeutic interventions (8). According to the Biomarkers Definitions Working Group (8), common applications of biomarkers include biomarkers which (I) can help diagnose a disease by identifying individuals with an abnormal biological process, (II) can classify disease severity, (III) can indicate prognosis or (IV) can predict or monitor response to therapy. Biomarkers have the potential to be utilized in several aspects of clinical care for patients with ASD, including promoting early diagnosis and selecting effective treatments.
This review will discuss emerging and promising biomarkers that could advance the diagnosis and treatment of ASD. The limitations of biomarkers and the current studies will be discussed along with insights into how biomarkers might be used in the future. Through this review, we hope the reader will gain a better understanding of the potential biomarkers that can be utilized in individuals with ASD and the complications and limitations of such biomarkers.
Methods
Search strategy
In order to identify studies of biomarkers in ASD, we conducted a review of the common medical literature databases PUBMED, Ovid Medline, Google Scholar, CINAHL, EmBase, Scopus, Cochrane and ERIC databases, from inception through June 2019. To identify studies, this review combined terms such as “biomarkers” and “autism”. Papers describing specific pathophysiology processes involved in ASD were also reviewed to determine if specific biomarkers had been developed for the specific research areas. Other filter terms such as “Human” or “Clinical Trials” were not uncommonly used. References in publications identified by the search were also reviewed to identify any relevant publications.
Study selection
Studies were included if they met specific criteria such as: (I) human randomized controlled trials, non-randomized trials, case studies and/or case series, and (II) reported a direct clinical biomarker as an outcome. Some animal studies were included if they supported the clinical studies discussed. After reviewing the identified studies, studies with promising findings were included in this review if they used well-known validated techniques to measure biomarkers and used well-accepted techniques to diagnose ASD. One of the goals of this review was to select publications that provided quantitative measurements of biomarker performance or applicability to the ASD population. However, some biomarkers without such quantitative evaluation were also included if there were multiple studies with positive results.
Rating study quality
A grade of recommendation (GOR) was given to allow the estimation of biomarker quality using the Oxford Centre for Evidence-Based Medicine scale. Those biomarkers that did not have clinical data to evaluate their use in clinical practice were not included in this evaluation. For each clinical study that described a biomarker, a level of evidence (LOE) ranging from 1 to 5 (see Table 1) was assigned (9). From these identified studies a GOR ranging from A (solid evidence) to D (limited, inconsistent or inconclusive evidence) was assigned (see Table 2).

Full table

Full table
Results and conclusions
From our review, it was found that biomarkers could be grouped into five different categories that are associated with the natural history of ASD. The potential promising biomarkers will be reviewed with reference to these categories:
- prenatal: starting from preconception through the gestation period, biomarkers have the potential to stratify pregnancies which may be high risk for the offspring developing ASD;
- pre-symptomatic: during the pre-symptomatic stages, biomarkers can identify high-risk populations to determine who may require further diagnostic testing, early intervention or increased surveillance;
- diagnostic: once symptoms are obvious, biomarkers can confirm the diagnosis;
- subgrouping: biomarkers can be used to divide individuals with ASD into biological subgroups;
- treatment: biomarkers can be used to select the most optimal therapy by predicting treatment response or measuring a physiological index of treatment response.
Our review of the literature also found that biomarkers fall into several major types; some examples are listed below:
- Behavioral (Eye Movements);
- Genetic (mRNA, miRNA, DNA, Methylation);
- Immune (Antibodies, Interleukin);
- Medical History (Family History, Pregnancy Complications);
- Metabolic (Methylation-Redox, Mitochondria, Amino Acids, Fatty Acids);
- Neuroimaging (Functional MRI, Structural MRI);
- Neurophysiology (EEG, MEG);
- Nutritional (Folate, Vitamin D, Vitamin B12).
Biomarkers in which associated quantitative data was reported are listed in Table 3. Quantitative measurements included the associated risk of developing ASD, the diagnostic accuracy, or the prevalence of the subgroup. The GOR for each biomarker is also outlined.
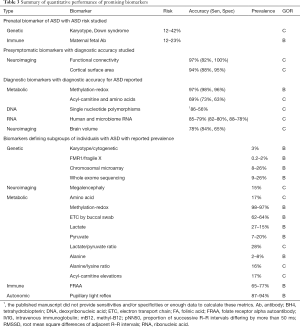
Full table
Prenatal biomarkers
Genetic
Most centers offer routine screening for genetic birth defects, most notably Down syndrome (DS) and other trisomy disorders. DS is the only commonly screened genetic disorder associated with ASD. Several studies have examined the association between DS and ASD. One population-based cohort of 41 children found a 42% prevalence using gold-standard diagnostic tools (10). Three studies have used the social communication questionnaire, a parent reported measure, to estimate the prevalence of ASD in DS. These studies, which examined 203, 108 and 507 individuals with DS, found a prevalence of 12% (11), 19% (12) and 38% (13), respectively. Since none of these trials are prospective, randomized or controlled they are LOE 4 trials, giving the overall C GOR. Despite the fact that the genetic disorder of DS can be detect, prenatal testing is very non-specific for the development of ASD in the child. Unfortunately, some countries promote prenatal genetic testing for ASD despite a lack of evidence for such a practice (14).
Immunologic
Immune activation during pregnancy increases the risk of ASD developing in the offspring. Both viral and bacterial maternal infections during pregnancy (15), particularly if they require hospitalization (16), increase the risk for offspring developing ASD. This evidence parallels the maternal immune activation (MIA) mouse model of ASD where an inflammatory event induced during pregnancy results in offspring with ASD-like behaviors (15). Although the mouse model has identified specific immunological mediators, including cytokines such as interleukin (IL)-6 and IL-17 (17), no human biomarker has been developed, so only the medical history of such an event can be used as a biomarker for identifying an increased risk of ASD.
During pregnancy, maternal immunoglobulin G (IgG) crosses the placenta, usually to protect the offspring during fetal life and early childhood. However, specific maternal IgG can target fetal brain tissue, causing a disruption in brain development. Early studies demonstrated that sera from mothers of children with ASD could induce behavioral abnormalities in both monkeys (18) and mice (19,20) when injected into the mothers. The first human controlled study (LOE 3b) identified that plasma from 12% of 61 mothers of children with ASD had two protein bands at 73 and 37 kDa that reacted with human fetal brain tissue (21). These proteins were not found in any of the 62 mothers with typically developing (TD) offspring nor any of the 40 mothers with children who had developmental delays but not ASD. These maternal IgG were found to target seven fetal proteins involved in neurodevelopment, including lactate dehydrogenase A and B, stress-induced phosphoprotein 1, collapsin response mediator proteins 1 and 2, cypin and Y-box binding protein (22). In a larger follow-up study (LOE 3b), 23% of 246 mothers of children with ASD demonstrated at least two of these seven antibodies while only 1% of 149 mothers of TD children demonstrated at least two of these seven antibodies. Children born from mothers with these antibodies had lower language (23,24) and cognitive ability (24), higher irritability (23), disrupted sleep-wake cycle (24) and large head circumference (24,25). With two LOE 3b studies, the GOR is B for maternal fetal brain antibodies.
Metabolic
ASD is associated with alternations in folate one-carbon metabolism (FOCM) and related pathways including methylation (26). Interestingly, maternal methylation abnormalities including DNA hypomethylation and abnormalities in metabolites integral to the methylation cycle, including plasma homocysteine, adenosine, and S-adenosylhomocysteine (SAM), were found in mothers who have offspring that developed ASD in two (LOE 3b) studies (27,28) providing a GOR B. However, these biomarkers were only able to determine whether the mother had high- or low-risk for having a child with ASD but not whether the offspring evidentially was diagnosed with ASD.
Nutritional
Folate during pregnancy has a complex relationship to the development of ASD in the offspring. Folate deficiency during pregnancy increases the risk of the offspring developing ASD (29) while folate supplement during pregnancy clearly decreases the risk of the offspring developing ASD (30). However, one study suggests that very high maternal folate or vitamin B12 blood concentrations at birth is associated with an increased risk of developing ASD and that both low and high folate supplementation increase the risk of developing ASD suggesting a U-shaped relationship between folate supplementation and ASD risk (31) (LOE 3b) but this notion has significant limitations when we consider the physiology of folate transport.
The transport of folate across the placenta may be blocked in mothers of children with ASD resulting in higher folate levels in the blood (32). Indeed, mothers of children with ASD have a higher prevalence of Folate Receptor Alpha Autoantibodies (FRAAs) (32) and a single nucleotide polymorphism (SNP) in the reduced folate carrier (RFC) (27), two mechanisms that disrupt folate transport across the placenta. The binding FRAA has also been associated with higher blood levels of vitamin B12 suggesting that it may inhibit transport of vitamin B12 into tissues (33). Furthermore, a recent animal model demonstrates that ASD-like behaviors develop in offspring in which folate transport across the placenta was blocked due to FRAAs (34). Despite this important data about folate and FRAAs prior to birth, no human studies have examined whether maternal FRAAs increase the risk of offspring developing ASD even though maternal FRAAs are linked to maternal subfertility (35) and neural tube defects in offspring (36-40). One case study (LOE 4) demonstrated that a FRAA positive woman with a history of multiple miscarriages and birth defects had a successful pregnancy after treatment targeting the FRAA abnormality (37).
Despite this evidence that folate transport across the placenta could be an explanation for changes in blood folate levels during pregnancy, other explanations for high blood folate levels are possible. For example, mothers who are at high risk for having a child with ASD may simply be ingesting higher doses of folate in order to protect their unborn child. Thus, the exact relationship between markers of folate metabolism and ASD risk is complex and it is too preliminary to provide a GOR at this time.
Other important nutritional biomarkers include vitamin D levels since lower 1st trimester vitamin D levels (41) (LOE 3b) and mid-gestation vitamin D deficiency (42) (LOE 2b) are associated with the severity of ASD behaviors in the offspring, and lifetime maternal vitamin D deficiency increases the risk of ASD in offspring (LOE 3b) (43). Thus, vitamin D has a GOR of B for predicting the diagnosis of ASD although the exact associated risk needs further study.
Given that treatment with specific folate compounds can decrease the risk of offspring developing ASD, this is clearly an important area to study (32). Nutritional interventions such as folate, vitamin D and vitamin B12 can have important health consequences, can potentially correct metabolic abnormalities noted above, and are generally regarded as safe. This area of physiology is potentially well suited for the development of safe interventions. Thus, the development of biomarkers for these nutritional abnormalities could be beneficial for preventing the development of ASD in the offspring or minimizing the severity of ASD.
Medical history
Several factors in the prenatal medical history are associated with an increased risk of ASD including advanced parental (44-46) and maternal age (45,46), birth order (44-46), and maternal prenatal factors including medications (29,44,46), smoking (29), bleeding (44), gestational diabetes (29,44,45), hypertension (44,45), proteinuria (44), pre-eclampsia (44), previous fetal loss (44), threatened abortion (45), antepartum hemorrhage (45), swelling (44), preeclampsia (45), fetal distress (45), and pre-pregnancy body mass index and gestational weight gain (47). During the perinatal period, factors associated with ASD such as caesarian delivery (45,46), prematurity (45,46) and breech presentation (45,46) have been reported. While there is no specific model that has been developed to predict the risk of a child developing ASD due to these risk factors, future studies could incorporate these factors into such calculations. However, several systematic reviews and meta-analyses (LOE 2a) (44-46) have been conducted on these factors providing a GOR B.
Fetal stress
A recent review discusses the growing research implicating maternal psychological stress, such as, partner abuse, bereavement and other psychosocial life stressors during the 2nd and 3rd trimester of pregnancy (48). These factors were revealed in several population-based cohort studies (LOE 2b) resulting in an GOR of B. One study did demonstrate that the risk of maternal bereavement on the offspring developing ASD was accounted for by other factors such as maternal psychiatric history, demonstrating the complicated interaction of many factors which could account for some of the reported effect. Still limitations of this research include the many different types of prenatal stress and the lack of one cohesive biomarker to measure such psychological stress.
Fetal toxicant exposure
Other studies have documented that prenatal exposure to environmental stressors such as toxicants can increase the risk of developing ASD although many of the studies used estimations of exposure rather than direct biomarker measurement (49). Three studies (LOE 2b) used urine biomarkers to measure maternal exposure to organophosphate and other toxicants during pregnancy resulting in a GOR of B (50-52). Particularly compelling as a biomarker is air pollution because air pollution is being measured with increasing precision and is publicly available information (53).
Post-natal pre-symptomatic biomarkers
Neuroimaging
Both functional and anatomic neuroimaging studies have demonstrated neuroimaging features that can predict the diagnosis of ASD in the early pre-symptomatic period. Using measures of functional connectivity derived from functional magnetic resonance imaging (MRI) scans at 6 months of age and machine learning, a diagnostic function was created which could predict the eventual diagnosis of ASD in 59 infants at high risk for developing ASD in a longitudinal non-randomized, non-controlled prospective study (LOE 2b) (54). By examining surface area obtained from MRI between 6 and 12 months of age, a deep-learning algorithm was able to predict the ASD diagnosis at 24 months in children with both high- and low-risk of developing ASD in a longitudinal non-randomized controlled prospective study (LOE 2b) (55). Although each individual technique receives a GOR of C, neuroimaging in general receives a GOR of B if both techniques are considered together.
Visual attention
Young children with ASD do not orient normally towards socially relevant point-light displays of biological motion (56) or other socially relevant stimuli (57). A prospective, longitudinal non-randomized controlled study measured visual attention in 110 infants with high- and low-risk for developing ASD from 2 to 24 months of age (LOE 2b) (58). For children that eventually developed ASD, visual attention to socially relevant stimuli started out normal at birth but then declined within the first 6-month life (58). Thus, abnormal early visual attention to social relevant stimuli is a promising early behavioral biomarker of ASD with a GOR C because of only one 2b study.
Neurophysiology
Resting state electroencephalography (EEG) was collected on 79 high- and low-risk infants at five time points from 6 to 24 months in a longitudinal, prospective non-randomized controlled study (LOE 2b). Multiscale entropy analysis was able to differentiate infants into risk groups with variable accuracy depending on gender, age and the diagnostic classifier used with the highest accuracy for boys at 9 months of age (59). Although this is a promising technique, this study did not look at ASD as an outcome measure.
Metabolic
One retrospective, population-based study (LOE 3b) assessed 3,258 3–5-year-old children with ASD and 6,838 controls without ASD and examined 36 analytes from newborn screening laboratory records. Five analytes [17-hydroxyprogesterone, free carnitine, isovalerylcarnitine (C5), octanoylcarnitine (C8), and phenylalanine/tyrosine ratio] were positively associated, and two [methylmalonylcarnitine (C4DC) and adipylcarnitine (C6DC)] were inversely associated with a later diagnosis of ASD (60). This provides a GOR of C for this large but non-validated metabolic analysis.
Autonomic nervous system (ANS)
Several studies have investigated imbalances in the ANS in children with ASD with some suggesting chronic hyperarousal (61,62) while others suggesting underarousal (63). In a prospective controlled study (LOE 2b) pupillary reactivity was studied in siblings of children with ASD as well as low-risk controls who were 9–10 months old (64). Children with an ASD diagnosis at 36 months of age demonstrated a relatively larger pupillary light reflex constriction as infants compared to the other groups with the magnitude of this response associated with symptom severity. With only one LOE 2b study the GOR is C.
Diagnostic biomarkers
Metabolic
ASD is associated with alternations in FOCM and related pathways include methylation and redox metabolism (26,65). These abnormalities result in oxidative damage to lipids, proteins and nucleic acids (26,65). Fisher Discriminant Analysis has been used to determine the diagnostic ability of biomarkers in these connected pathways. In the first prospective, controlled non-randomized cross-sectional study (LOE 2b), 83 individuals with ASD were differentiated from 76 healthy controls with 97% accuracy (66). In a separate validation study using prospective non-randomized data from 154 individuals with ASD studied in three clinical trials (LOE 2b), a 97% sensitivity was found using a similar classification model as the first study (67). Given two LOE 2b studies, the GOR is B.
In a prospective, controlled non-randomized study (LOE 2b), amino acids and acyl-carnitines derived from dried blood spots correctly classify ASD and control participants with 69% accuracy (68). Accuracy was 69% in a validation sample and the sensitivity was 68%. Given only one LOE 2b study, the GOR is C.
Proteomics
Techniques for measuring specific proteins (targeted) and novel proteins (untargeted) have been applied to blood, urine and saliva samples with inconsistent results across studies. Untargeted analysis of serum has demonstrated involvement of complement (69,70), cholesterol metabolism (71) and apoptosis proteins (69). Using targeted techniques, analysis of serum has identified inflammation and growth factor proteins (72). One (n=60) case-control age and gender matched, non-randomized study (LOE 2b) using isobaric tags for relative and absolute quantitation (iTRAQ) found that a combination of five proteins (C3, C5, GC, ITGA2B, and TLN1) could distinguish those with and without ASD with an impressive area under the curve of 0.982 (73). With only one clinical outcome study, the GOR is a C.
SNPs
A diagnostic classifier based of 237 SNPs was applied to three large ASD genetic databases in a retrospective, non-randomized design (LOE 2b). Individuals with ASD were distinguished from controls with an accuracy varying from 56% to 86%, depending on the ethnic similarity between the training and validation datasets (74). The sensitivity of the classifier to the ethnicity of the dataset demonstrates the potential complications of using genetic information.
Ribonucleic acid (RNA)
Cellular physiology is controlled by how genes are expressed (epigenetics), which is different from the actual coding sequence of a gene. Thus, several studies have measured various types of RNA as a biomarker to investigate cellular physiology. One intriguing, multicenter, non-randomized controlled prospective study (LOE 2b) examined both the human and microbiome RNA obtained from the oral cavity. RNA was subdivided into five types: microRNA, piwi-interacting RNA, non-coding RNA, ribosomal RNA and microbial RNA. Using machine learning and hold-out validation, 32 diagnostic features obtained a 79% accuracy in a training data (n=372). The smaller (n=84) new validation dataset performed slightly better with an accuracy of 85% (75).
Neurophysiology
The Autism Biomarkers Consortium for Clinical Trials (ABC-CT) (76) project is aimed to develop a neurophysiological biomarker of ASD. This project focuses on the N170 response to face stimuli (77). Delayed and lower amplitude N170 responses have been documented in children (78,79), adolescents (78,80) and adults (80) with ASD and children and adults with Asperger’s syndrome (81). A systematic-review and meta-analysis of the N170 (77) (LOE 2a) suggests that the latency to face stimuli are indeed delayed in individuals with ASD with the M170 amplitude affected by several factors such as age and cognitive ability. Thus, the M170 has a GOR of B as a biomarker, but its ability to classify ASD from controls has not been studied; therefore, its performance as a diagnostic biomarker for ASD is not known.
Other studies have associated neurophysiological alterations with ASD. Suppression of the mu rhythm, which is thought to reflect dysfunction in mirror neurons, is reduced in high-functioning adults with ASD while performing (82,83) or observing (83) a motor task, and in children with ASD while observing motor tasks (84) in three small controlled studies (LOE 2b; GOR B). Individuals with ASD demonstrate atypical gamma-band oscillations when viewing complex visual stimuli. The relationship between the spatial frequency of Gabor patches and gamma-band power was reduced in or near the striate or extrastriate cortex in individuals with ASD (85). However, the homogeneity of the orientation of small line-like elements did not influence the gamma-band response in high-functioning adults with ASD (86). In another study, viewing illusory Kanizsa shapes resulted in differences in the timing of gamma-band peaks between TD and ASD adolescents (87) and children (88). Finally, impaired perception of Mooney faces was related to a reduced amplitude and phase locking of gamma-band activity in adults with ASD (89). Thus, five small controlled studies (LOE 2b; GOR B) suggest that gamma-band activity could be a promising biomarker for ASD.
Magnetoencephalography (MEG) has been used to study the response of the auditory system in children with ASD. Three small controlled studies (LOE 2b) have identified delayed M100 and auditory mismatch field in adults (90) and children (91) with ASD and have correlated changes in these neurophysiological parameters with ASD severity (91,92) (GOR B). A large controlled study (LOE 2b) has pointed to abnormal oscillatory activity during the pre-stimulus period in the superior temporal gyrus during an auditory task (93) (GOR C).
Neuroimaging
Following research on anatomic brain development, one rather large (59 ASD, 77 controls) prospective, non-randomized, controlled study examined extra-axial fluid using MRI (LOE 2b). Extra-axial fluid volume has associated with sleep disturbance and lower verbal ability (94). Machine learning data analysis of extra-axial and brain volumetric data, along with subject age and gender, resulted in a diagnostic predictive function with 78% accuracy (GOR C).
Biomarkers identifying subgroups of ASD
Genetic
ASD is associated with a 70–90% concordance rate for monozygotic twins and the risk of having a second child with ASD is increased 25-fold for families who already have a child with ASD (95). This high heritability has driven the notion that genetic factors are a major contributor to the etiology of ASD (96,97). Some studies estimate that up to 40% of cases of ASD may have a genetic underpinning (98,99). However, this high rate of genetic disorders was not confirmed in at least one large-scale study (99). Nevertheless, genetic testing is standard-of-care for children with ASD because identifying a genetic etiology might help with prescribing treatments and prognosis.
The most basic genetic biomarker is the karyotype that examines the gross chromosomal structure. Cytogenetic abnormalities are reported in 3% of individuals with ASD in two studies (both LOE 2b) (100,101). Karyotype testing has largely been eclipsed by the chromosomal microarray (CMA) which can detect duplications or deletions of small portions of chromosomes, usually down to 15,000 bp (102). Several different specific copy number variations (CNVs) have been associated with ASD (102) with an estimation of the prevalence of these CNVs varying between 8% (99) (LOE 2b) and 26% (103) (LOE 2b). As a complement to the CMA, whole exome sequencing has revolutionized the ability to identify single gene disorders. Using this technique, 9% (99) (LOE 2b) to 30% (104) of children with ASD have been estimated to have a single gene disorder.
An important genetic disorder that is not identified with the aforementioned techniques is Fragile X (98). This disorder expresses itself when there are more than 200 CGG repeats in the FMR1 gene on the X-chromosome. Testing for Fragile X is recommended as standard-of-care for ASD (98). Fragile X Syndrome is reported in 0.2% (LOE 3b) (105) to 2% (LOE 2b) (100) of individuals with ASD.
Genetic testing continues to evolve with some geneticists performing whole genome sequencing to identify disorders with involvement of non-coding regions of the genome and examining the complex influences of polygenetic factors. One recent retrospective study of 18,381 cases of ASD and 27,969 controls (LOE 3b) reported 13 genes associated with ASD which were outside the known genome (106). Other studies have examined the patterns of SNPs associated with ASD. Of course, although the various genetic technique may identify different abnormalities, these abnormalities are not mutually exclusive, and evidence suggests that those with CNV also harbor other genetic mutations. Several reviews have discussed the complicated landscape of potential genetic causes of ASD (102).
Neuroimaging
One of the neurophenotypes identified as part of the Autism Phenome Project was boys with megalencephaly, a brain that was disproportionately large as compared to body size. The estimated 15% of boys with ASD who have this phenotype had more severe disabilities and had a poorer prognosis (107) (LOE 2b).
Metabolic
Amino acids
The Children’s Autism Metabolome Project (CAMP) was a large prospective, controlled non-randomized multisite study aimed at discovering biomarkers of ASD and compared 516 children with ASD to 164 age-matched TD controls (LOE 2b) (108). This study identified a subgroup with dysregulated branched chain amino acid metabolism found in 17% of children with ASD with a 96% specificity. The CAMP method only has an accuracy of 36% and specificity of 17% for the diagnosis of ASD, so it can only identify a subgroup of children with ASD. Thus, it is not useful on its own as a diagnostic biomarker of ASD. However, the exciting aspect of this biomarker is that branched chain amino acid supplementation is a potential treatment for ASD. However, the CAMP biomarker cannot be considered a treatment biomarker since this treatment has not been studied in ASD. In the genetic form of this disorder, supplementation with branched chain amino acids improved symptoms in the mouse model but not in the human patients (109).
Methylation-redox biomarkers
ASD is associated with alternations in FOCM related to methylation and redox metabolism abnormalities (26,65). Dr. James originally described this unique ASD endophenotype (26) in three prospective, non-randomized case-control studies (LOE 2b) (26,110,111). Abnormalities in redox metabolism as measured by alterations in glutathione redox status were also verified in case-control studies (LOE 2b) utilizing primary immune cells (112), lymphoblastoid cell lines (113) and post-mortem brain tissue (114). A meta-analysis and systematic review (LOE 2a) verified abnormal redox biomarkers related to ASD including abnormalities in glutathione, methionine and cysteine across repeated studies from several different laboratories (115). As described above, Fisher Discriminant Analysis found that biomarkers of methylation and redox metabolism and oxidative damage identified a subgroup of 98% of children with ASD in one study (LOE 2b) (66) and 97% of children with ASD in another study (LOE 2b) (67). Therefore methylation-redox markers are GOR B.
Mitochondrial biomarkers
Approximately 5% of children with ASD exhibit mitochondrial disease (116) while many more, perhaps between 30–80% exhibit some degree of mitochondrial dysfunction (117). Many biomarkers of mitochondrial dysfunction have been used in ASD, as recently reviewed (117). Using the buccal swab technique to measure electron transport chain (ETC) complex activity, abnormal mitochondrial function was identified in 64% of children with ASD in one prospective study (LOE 2b) (118) and 62% in one retrospective study (LOE 3b) (119). A previous meta-analysis estimated the prevalence of lactate (31.1%, 6 LOE 3b studies), pyruvate (13.6%, 2 LOE 3b studies) and lactate-to-pyruvate ratio (27.6%, 1 LOE 3b study) abnormalities (116). The prevalence of alanine abnormalities was reported in the meta-analysis (8.3%, LOE 2a) (116) and in a retrospective analysis of an ASD clinic (120) (1.7%, LOE 3b). The prevalence of an abnormal alanine-to-lysine ratio was reviewed in a retrospective analysis of an ASD clinic (120) (15.9%, LOE 3b). The prevalence of a unique pattern of elevations in short and long but not medium chain acyl-carnitines was discovered in a retrospective analysis of an ASD clinic (LOE 3b) as 24% (120) but refined in a larger retrospective analysis (LOE 3b) of the same clinic as 17% (121).
Other standard blood markers such as creatine kinase, ammonia and aspartate aminotransferase (AST) are commonly used biomarkers for mitochondrial dysfunction in ASD but the studies are too small to estimate the prevalence of any of these specific biomarkers being abnormal in ASD (116). Others have shown that ASD and TD children can be differentiated by novel biomarkers of mitochondrial dysfunction such as lactate dehydrogenase (122), lactate oxidase, pyruvate kinase and hexokinase (123), Na+/K+ ATPase (124,125), Caspase-3 (126) and Caspase-7 (122). Again, these studies are not consistent in biomarkers used to differentiate the groups. Thus, while these studies do provide evidence for biomarkers of mitochondrial dysfunction being useful in identifying subgroups of individuals with ASD, the inconsistency prevents a clear prevalence to be estimated and it is unclear how to most appropriately combine these studies to judge a GOR.
The first study which examined possible carnitine deficiency used adult normative values as controls so the accuracy of the study conclusions has been questioned (127). However, other studies have verified these abnormalities in carnitine levels, particularly in individuals with ASD and GI abnormalities (128). Additionally, a genetic abnormality in carnitine metabolism has been linked to ASD (129) and primary carnitine deficiency was reported to cause ASD in one case report (130). Three clinical trials have reported that carnitine supplementation improves symptoms in ASD (131-133). Thus, the importance of carnitine is being recognized in ASD and its deficiency is being acknowledged as a potential biomarker of a metabolic subgroup of ASD. However, given the inconsistency in the approaches of the studies on carnitine it is difficult to estimate a prevalence or a GOR.
Immune
In an open-label clinical study (LOE 2b), 75% of the participants had at least one FRAA (134) while 65% of the participants had at least one FRAA in a prospective, double-blind placebo controlled (DBPC) clinical trial (LOE 1b) (135). The significance of the FRAA as a biomarker of an ASD subgroup with unique characteristics has been reported (33).
Alternations in gut permeability
Alterations in intestinal barrier integrity has been identified in patients with ASD in several studies (136) with one study demonstrating reduced expression of barrier-forming intestine “tight junction” components in 75% of patients with ASD (137). Zonulin regulates tight junctions between enterocytes and is a physiological modulator controlling intestinal permeability. This protein was increased in patients with ASD compared with healthy controls and was correlated with measures of ASD severity (138) (LOE 3b). Zonulin may be a promising biomarker for a subgroup of children with ASD with GI problems stemming from altered intestinal integrity with a GOR of C. However, it should also be mentioned that not all studies support that notion of increased gut permeability (139).
Autonomic nervous system
As previously mentioned, ANS abnormalities have been noted in individuals with ASD. Many studies have concentrated on cardiac measures such as heart rate variability (61) while other studies have focused on pupillary response (140) as measures of ANS biomarkers. Heart rate variability has been studied in adults (141-144) and children (63,145-147) with ASD as well as their caregivers (148-150) and has been associated with GI symptoms (146) and used as an outcome measure in clinical treatments studies (147,151,152). Given the multiple LOE 3b and 2b studies, heart rate variability has a GOR of B. Pupillary light reflex was studied in four prospective nonrandomized controlled studies of children with ASD (LOE 2b) (153) with one study linked these abnormalities to the severity of atypical sensory behaviors (154) and heart rate variability (155), and one study demonstrating the ability of pupillary light reflex parameters to accurately select a subgroup of 86.7–93.7% of the ASD participants with a 92.5–96.7% accuracy (156). A recent review discusses some of the limitations of these measures and the questions that need to be addressed in order to develop ANS biomarkers further (157). The Guidelines for Reporting Articles on Psychiatry and Heart rate variability (GRAPH) efforts has been launched to standardize reporting of at least heart rate variability so consistent data can be provided across studies for valid comparisons (158).
Biomarkers that predict treatment response
Three metabolic treatments have biomarkers that correlate with treatment response (Table 4). Core ASD symptoms improve with carnitine supplementation with one study demonstrating that the increase in blood carnitine levels were related to improvements in physical and cognitive abilities (LOE 2b) (131). Two methylcobalamin treatment studies, one open label (LOE 4) and one DBPC (LOE 2b) demonstrate a correspondence between changes in methylation and redox metabolites and functional abilities (159) and ASD symptoms (160,161). Lastly, biomarkers of tetrahydrobiopterin metabolism correlated with language, behavior and functional improvements following tetrahydrobiopterin treatment in an open-label study (LOE 4) (161,162).
Two studies demonstrate that autoantibodies can predict response to specific treatment (Table 4). Pretreatment FRAA titers predicted response to leucovorin calcium in children with language delay in a DBPC study (LOE 2b) (135). For patients with ASD and autoimmune encephalopathy, specific brain autoantibodies, particularly those to the D2L receptor and tubulin, predicted response to treatment with intravenous immunoglobulin in an open-label study (LOE 4) (163).
Medications that modulate the ANS, particularly the beta-adrenergic antagonist propranolol, have emerged as potential effective treatments for the core features of ASD (164). In a prospective, DBPC study (LOE 2b) of the effect of propranolol on verbal problem solving in adults and adolescents with ASD, two ANS biomarkers, the root mean square differences of adjacent R–R intervals (RMSSD) and the proportion of successive R–R intervals differing by more than 50 ms (pNN50), predicted treatment response (165) (Table 4).
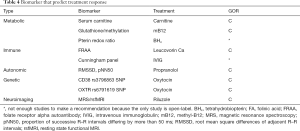
Full table
Treatment with the hormone oxytocin is undergoing particularly active study (164). Two SNPs, one on the CD38 gene which codes for the transmembrane protein involved in oxytocin secretion (166) and another on the OXTR gene which codes with the oxytocin receptor (167) have been found to predict response to oxytocin in clinical studies (both LOE 2b).
In a medium size (n=37) DBPC cross-over study (LOE 2b) researcher used magnetic resonance spectroscopy and resting state functional MRI to demonstrate that riluzole had the predicted response on improving the cortical excitatory to inhibitory balance and improved functional connectivity in the frontal lobes (168).
Limitations of biomarker studies
One of the major limitations in biomarkers studies is the comparison groups used. Many studies compare individuals with ASD to TD unrelated controls. Although scientifically valid, this comparison has some serious limitations when considering the use of biomarkers in the real clinical world. Specifically, in the clinic, the question that needs to be answered is not whether a completely normally developing child has ASD but rather whether a child with developmental delays or an unusual behavior may have a diagnosis of ASD. Likewise, another important question is whether a sibling of a child who is already diagnosed with ASD will develop ASD since the sibling is at higher risk. Thus, although TD unrelated controls may be adequate for initially developing biomarkers, validation studies need to use more rigorous and clinically relevant controls.
The other important limitation of many biomarkers, particularly diagnostic biomarkers, is their biological validity. For example, the ability of FRAAs to predict clinical response to treatment with leucovorin is consistent with its proposed scientific physiological mechanism of action. However, if a biomarker does not represent a core biological process of the disease, there is a danger that the biomarker represents an epiphenomenon of the disease process. Thus, even if the biomarker reliably and consistently differentiates groups of patients, the information provided about the underlying disease process may be limited.
There are many emerging biomarkers for ASD, but biomarkers need to be used in the correct context. For example, although FRAAs may be very useful in guiding therapy for those already diagnosed with ASD, FRAAs are probably not be a good test to establish a diagnosis of ASD, since they are only positive in 50–70% of children with ASD.
The promise of the use of biomarkers in the future
Even though diagnostic ASD behavior may not be clearly defined until late in the second year of life, the neuropathology associated with ASD may begin prenatally in some cases. Thus, there is the potential for biomarkers that aid in identifying individuals at risk for developing ASD well before diagnostic behaviors are clearly established. Potential biomarkers have been identified but the predictive power of most have not been studied. The most promising prenatal biomarker is maternal fetal brain autoantibodies that appear to have good specificity for an offspring developing ASD. However, while this antibody panel may appear highly translatable as a clinical tool, the lack of any proven associated treatment has reduced enthusiasm to develop a widely-used commercial test (169). Postnatal presymptomatic neuroimaging biomarkers are promising but further studies are needed to validate their performance.
Several biomarkers are being developed as diagnostic tools. Many of the studies remain preliminary and most of the biomarkers do not have adequate performance. Given the significant heterogeneity in the etiology of ASD, large samples of individuals with ASD as well as large, carefully selected clinically relevant control groups will be needed to validate such biomarkers. It is likely that the performance of these biomarkers will fail to operate adequately if considered in isolation and used as a general screening tool. Thus, any diagnostic biomarker developed needs to measure biological processes and most likely will need to be used in conjunction with other behavioral tests and clinically relevant information to perform adequately.
Research supports the notion that the etiology of ASD arises from a complex interaction between genetic susceptibility and environmental exposures (49,170). Thus, an exciting possibility of biomarker use in ASD is in prevention by identifying those at risk during the prenatal or pre-symptomatic period so abnormal biological processes can be identified early, and potential triggers can be avoided, or biological abnormalities can be addressed. Although environmental exposures have been identified (prenatal stress, toxicants), there are no studies examining biomarkers that measure these environmental risks nor have the biological processes that may stratify risk been carefully studied. Thus, biomarkers that can stratify susceptibility risk in order to avoid exposure to environmental triggers might help avoid the development of ASD in the first place. Such studies could identify vulnerable populations that could be protected from environmental triggers.
Biomarkers may also be extremely helpful in explaining the significant heterogeneity associated with ASD (171). The underlying pathophysiology that drives ASD symptoms may be very different patient-to-patient. For example, some individuals have a well-established genetic syndrome while others demonstrate other non-genetic physiological abnormalities. Biomarkers that can subdivide patients into clear etiological subgroups may be very helpful for prescribing optimal treatments and predicting prognosis. This seems to be the most well-developed aspect of biomarkers, but studies remain in the beginning stages.
Many individuals with ASD are medically complex with multiple co-morbid medical conditions. Unfortunately, these medical conditions are not always obvious. For example, many GI disorders can lead to symptoms that are considered core ASD symptoms such as repetitive behaviors; thus, such symptoms are commonly not recognized as indicative of GI distress (172). Biomarkers that can identify these medical conditions can help target those who need specific evaluation and treatment. There is only one study examining the promising biomarker Zonulin that has the potential to identify those with GI disorders. However, several biomarkers have been developed to identify mitochondrial dysfunction and methylation-redox abnormalities. These biomarkers may be extremely helpful because metabolic disorders are potentially treatable conditions. Additionally, markers of ANS dysfunction may be very promising as such abnormalities may drive disruptive behaviors and have potential treatments.
Individual response to ASD treatment varies widely, so biomarkers that can predict treatment response could be very helpful for optimizing individualized treatment plans, leading to a personalized precision medicine approach (171). Although several biomarkers that can predict treatment response have been described, the studies validating them are in the early stages. Many of these biomarkers have also been studied for selecting biological subgroups, which enhances their validity and clinical applicability. Subgroup and treatment biomarkers may be some of the most powerful as few guidelines are available for selecting the most appropriate treatment. Given that there are current few outcome measures to objectively monitor treatment effectiveness, treatment biomarkers may be the most promising and have the most impact.
In the future we may learn to use biomarkers together in an integrated way and learn to map them onto specific important symptoms or therapeutic actions. As an example, the National Institute of Mental Health has developed the Research Domain Criteria for studying mental health (173). This initiative divides human mental functioning into five specific domains which contains specific behavioral or emotional constructs. Various elements from genes to molecules to behaviors are mapped onto each construct in order to better understand the factors that drive the construct as well as the instruments that can measure the construct. In this way, elements of mental disease are systematically divided into understandable parts. As symptoms of ASD are mapped onto specific biological processes and biomarkers of such processes, insight into disease processes and potential therapeutics should emerge.
Conclusions
Promising biomarkers identified in this review fall into five types (Figure 1). Although there are many potential biomarkers for predicting the risk of developing ASD before birth, many of these biomarkers have not been systematically studied to generate estimations of the associated risk of developing ASD after birth. Several pre-symptomatic biomarkers are being developed but only two have an estimation of diagnostic accuracy based on very preliminary studies. Several diagnostic biomarkers are promising but like the pre-symptomatic biomarkers, the studies remain very preliminary. For example, one of the major efforts to develop a diagnostic biomarker, ABC-CT, does not have a study to estimate its diagnostic performance. Subgrouping biomarkers appear the most robust in regard to the number of biomarkers and the number of studies conducted that estimate the size of the subgroup. Biomarkers that predict treatment response are developing, but data supporting them are preliminary. Overall, the evidence for biomarkers in ASD is preliminary but promising.

Acknowledgments
None.
Footnote
Conflicts of Interest: Dr. Frye is funded by the National Institutes of Health, Department of Defense, and Autism Speaks and receives support from the The Turnabout for Autism and Autism Research Institute foundations and Zynerba Pharmaceuticals. The author is on the advisory boards of Iliad Neurosciences, NeuroNeeds and Cox Biosciences. Dr. Rossignol is on the advisory board of NeuroNeeds.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Christensen DL, Braun KVN, Baio J, et al. Prevalence and Characteristics of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2012. MMWR Surveill Summ 2018;65:1-23. [Crossref] [PubMed]
- Xu G, Strathearn L, Liu B, et al. Prevalence and Treatment Patterns of Autism Spectrum Disorder in the United States, 2016. JAMA Pediatr 2019;173:153-9. [Crossref] [PubMed]
- Rogge N, Janssen J. The Economic Costs of Autism Spectrum Disorder: A Literature Review. J Autism Dev Disord 2019;49:2873-900. [Crossref] [PubMed]
- Leigh JP, Du J. Brief Report: Forecasting the Economic Burden of Autism in 2015 and 2025 in the United States. J Autism Dev Disord 2015;45:4135-9. [Crossref] [PubMed]
- Brown CC, Tilford JM, Payakachat N, et al. Measuring Health Spillover Effects in Caregivers of Children with Autism Spectrum Disorder: A Comparison of the EQ-5D-3L and SF-6D. Pharmacoeconomics 2019;37:609-20. [Crossref] [PubMed]
- Hartley SL, Papp LM, Bolt D. Spillover of Marital Interactions and Parenting Stress in Families of Children With Autism Spectrum Disorder. J Clin Child Adolesc Psychol 2018;47:S88-99. [Crossref] [PubMed]
- Tilford JM, Payakachat N, Kuhlthau KA, et al. Treatment for Sleep Problems in Children with Autism and Caregiver Spillover Effects. J Autism Dev Disord 2015;45:3613-23. [Crossref] [PubMed]
- Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther 2001;69:89-95. [Crossref] [PubMed]
- Howick J, Chalmers J, Glasziou P, et al. The 2011 Oxford CEBM Levels of Evidence. Oxford Centre for Evidence-Based Medicine. 2011. Available online: https://www.cebm.net/index.aspx?o=5653
- Oxelgren UW, Myrelid A, Anneren G, et al. Prevalence of autism and attention-deficit-hyperactivity disorder in Down syndrome: a population-based study. Dev Med Child Neurol 2017;59:276-83. [Crossref] [PubMed]
- Channell MM, Hahn LJ, Rosser TC, et al. Characteristics Associated with Autism Spectrum Disorder Risk in Individuals with Down Syndrome. J Autism Dev Disord 2019;49:3543-56. [Crossref] [PubMed]
- Moss J, Richards C, Nelson L, et al. Prevalence of autism spectrum disorder symptomatology and related behavioural characteristics in individuals with Down syndrome. Autism 2013;17:390-404. [Crossref] [PubMed]
- Warner G, Moss J, Smith P, et al. Autism characteristics and behavioural disturbances in ~ 500 children with Down's syndrome in England and Wales. Autism Res 2014;7:433-41. [Crossref] [PubMed]
- Zhao S, Wang CH, Huang TY, et al. A qualitative study exploring the attitudes toward prenatal genetic testing for autism spectrum disorders among parents of affected children in Taiwan Research in Autism Spectrum Disorders 2018;48:36-43. [Crossref]
- Patterson PH. Maternal infection and immune involvement in autism. Trends Mol Med 2011;17:389-94. [Crossref] [PubMed]
- Jiang HY, Xu LL, Shao L, et al. Maternal infection during pregnancy and risk of autism spectrum disorders: A systematic review and meta-analysis. Brain Behav Immun 2016;58:165-72. [Crossref] [PubMed]
- Brimberg L, Mader S, Jeganathan V, et al. Caspr2-reactive antibody cloned from a mother of an ASD child mediates an ASD-like phenotype in mice. Mol Psychiatry 2016;21:1663-71. [Crossref] [PubMed]
- Martin LA, Ashwood P, Braunschweig D, et al. Stereotypies and hyperactivity in rhesus monkeys exposed to IgG from mothers of children with autism. Brain Behav Immun 2008;22:806-16. [Crossref] [PubMed]
- Singer HS, Morris C, Gause C, et al. Prenatal exposure to antibodies from mothers of children with autism produces neurobehavioral alterations: A pregnant dam mouse model. J Neuroimmunol 2009;211:39-48. [Crossref] [PubMed]
- Dalton P, Deacon R, Blamire A, et al. Maternal neuronal antibodies associated with autism and a language disorder. Ann Neurol 2003;53:533-7. [Crossref] [PubMed]
- Braunschweig D, Ashwood P, Krakowiak P, et al. Autism: maternally derived antibodies specific for fetal brain proteins. Neurotoxicology 2008;29:226-31. [PubMed]
- Braunschweig D, Krakowiak P, Duncanson P, et al. Autism-specific maternal autoantibodies recognize critical proteins in developing brain. Transl Psychiatry 2013;3:e277. [Crossref] [PubMed]
- Braunschweig D, Duncanson P, Boyce R, et al. Behavioral correlates of maternal antibody status among children with autism. J Autism Dev Disord 2012;42:1435-45. [Crossref] [PubMed]
- Piras IS, Haapanen L, Napolioni V, et al. Anti-brain antibodies are associated with more severe cognitive and behavioral profiles in Italian children with Autism Spectrum Disorder. Brain Behav Immun 2014;38:91-9. [Crossref] [PubMed]
- Nordahl CW, Braunschweig D, Iosif AM, et al. Maternal autoantibodies are associated with abnormal brain enlargement in a subgroup of children with autism spectrum disorder. Brain Behav Immun 2013;30:61-5. [Crossref] [PubMed]
- James SJ, Melnyk S, Jernigan S, et al. Metabolic endophenotype and related genotypes are associated with oxidative stress in children with autism. Am J Med Genet B Neuropsychiatr Genet 2006;141B:947-56. [Crossref] [PubMed]
- James SJ, Melnyk S, Jernigan S, et al. A functional polymorphism in the reduced folate carrier gene and DNA hypomethylation in mothers of children with autism. Am J Med Genet B Neuropsychiatr Genet 2010;153B:1209-20. [PubMed]
- Hollowood K, Melnyk S, Pavliv O, et al. Maternal metabolic profile predicts high or low risk of an autism pregnancy outcome. Res Autism Spectr Disord 2018;56:72-82. [Crossref] [PubMed]
- Ornoy A, Weinstein-Fudim L, Ergaz Z. Prenatal factors associated with autism spectrum disorder (ASD). Reprod Toxicol 2015;56:155-69. [Crossref] [PubMed]
- Levine SZ, Kodesh A, Viktorin A, et al. Association of Maternal Use of Folic Acid and Multivitamin Supplements in the Periods Before and During Pregnancy With the Risk of Autism Spectrum Disorder in Offspring. JAMA Psychiatry 2018;75:176-84. [Crossref] [PubMed]
- Raghavan R, Riley AW, Volk H, et al. Maternal Multivitamin Intake, Plasma Folate and Vitamin B12 Levels and Autism Spectrum Disorder Risk in Offspring. Paediatr Perinat Epidemiol 2018;32:100-11. [Crossref] [PubMed]
- Frye RE, Slattery JC, Quadros EV. Folate metabolism abnormalities in autism: potential biomarkers. Biomark Med 2017;11:687-99. [Crossref] [PubMed]
- Frye RE, Delhey L, Slattery J, et al. Blocking and Binding Folate Receptor Alpha Autoantibodies Identify Novel Autism Spectrum Disorder Subgroups. Front Neurosci 2016;10:80. [Crossref] [PubMed]
- Sequeira JM, Desai A, Berrocal-Zaragoza MI, et al. Exposure to Folate Receptor Alpha Antibodies during Gestation and Weaning Leads to Severe Behavioral Deficits in Rats: A Pilot Study. PLoS One 2016;11:e0152249. [Crossref] [PubMed]
- Berrocal-Zaragoza MI, Fernandez-Ballart JD, Murphy MM, et al. Association between blocking folate receptor autoantibodies and subfertility. Fertil Steril 2009;91:1518-21. [Crossref] [PubMed]
- Yang N, Wang L, Finnell RH, et al. Levels of folate receptor autoantibodies in maternal and cord blood and risk of neural tube defects in a Chinese population. Birth Defects Res A Clin Mol Teratol 2016;106:685-95. [Crossref] [PubMed]
- Shapira I, Sequeira JM, Quadros EV. Folate receptor autoantibodies in pregnancy related complications. Birth Defects Res A Clin Mol Teratol 2015;103:1028-30. [Crossref] [PubMed]
- Boyles AL, Ballard JL, Gorman EB, et al. Association between inhibited binding of folic acid to folate receptor alpha in maternal serum and folate-related birth defects in Norway. Hum Reprod 2011;26:2232-8. [Crossref] [PubMed]
- Cabrera RM, Shaw GM, Ballard JL, et al. Autoantibodies to folate receptor during pregnancy and neural tube defect risk. J Reprod Immunol 2008;79:85-92. [Crossref] [PubMed]
- Rothenberg SP, da Costa MP, Sequeira JM, et al. Autoantibodies against folate receptors in women with a pregnancy complicated by a neural-tube defect. N Engl J Med 2004;350:134-42. [Crossref] [PubMed]
- Chen J, Xin K, Wei J, et al. Lower maternal serum 25(OH) D in first trimester associated with higher autism risk in Chinese offspring. J Psychosom Res 2016;89:98-101. [Crossref] [PubMed]
- Vinkhuyzen AAE, Eyles DW, Burne THJ, et al. Gestational vitamin D deficiency and autism spectrum disorder. BJPsych Open 2017;3:85-90. [Crossref] [PubMed]
- Magnusson C, Lundberg M, Lee BK, et al. Maternal vitamin D deficiency and the risk of autism spectrum disorders: population-based study. BJPsych Open 2016;2:170-2. [Crossref] [PubMed]
- Gardener H, Spiegelman D, Buka SL. Prenatal risk factors for autism: comprehensive meta-analysis. Br J Psychiatry 2009;195:7-14. [Crossref] [PubMed]
- Wang C, Geng H, Liu W, et al. Prenatal, perinatal, and postnatal factors associated with autism: A meta-analysis. Medicine (Baltimore) 2017;96:e6696. [Crossref] [PubMed]
- Guinchat V, Thorsen P, Laurent C, et al. Pre-, peri- and neonatal risk factors for autism. Acta Obstet Gynecol Scand 2012;91:287-300. [Crossref] [PubMed]
- Shen Y, Dong H, Lu X, et al. Associations among maternal pre-pregnancy body mass index, gestational weight gain and risk of autism in the Han Chinese population. BMC Psychiatry 2018;18:11. [Crossref] [PubMed]
- Beversdorf DQ, Stevens HE, Jones KL. Prenatal Stress, Maternal Immune Dysregulation, and Their Association With Autism Spectrum Disorders. Curr Psychiatry Rep 2018;20:76. [Crossref] [PubMed]
- Rossignol DA, Genuis SJ, Frye RE. Environmental toxicants and autism spectrum disorders: a systematic review. Transl Psychiatry 2014;4:e360. [Crossref] [PubMed]
- Eskenazi B, Marks AR, Bradman A, et al. Organophosphate pesticide exposure and neurodevelopment in young Mexican-American children. Environ Health Perspect 2007;115:792-8. [Crossref] [PubMed]
- Cheslack-Postava K, Rantakokko PV, Hinkka-Yli-Salomaki S, et al. Maternal serum persistent organic pollutants in the Finnish Prenatal Study of Autism: A pilot study. Neurotoxicol Teratol 2013;38:1-5. [Crossref] [PubMed]
- Miodovnik A, Engel SM, Zhu C, et al. Endocrine disruptors and childhood social impairment. Neurotoxicology 2011;32:261-7. [Crossref] [PubMed]
- Kaufman JA, Wright JM, Rice G, et al. Ambient ozone and fine particulate matter exposures and autism spectrum disorder in metropolitan Cincinnati, Ohio. Environ Res 2019;171:218-27. [Crossref] [PubMed]
- Emerson RW, Adams C, Nishino T, et al. Functional neuroimaging of high-risk 6-month-old infants predicts a diagnosis of autism at 24 months of age. Sci Transl Med 2017. [Crossref] [PubMed]
- Hazlett HC, Gu H, Munsell BC, et al. Early brain development in infants at high risk for autism spectrum disorder. Nature 2017;542:348-51. [Crossref] [PubMed]
- Klin A, Lin DJ, Gorrindo P, et al. Two-year-olds with autism orient to non-social contingencies rather than biological motion. Nature 2009;459:257-61. [Crossref] [PubMed]
- Klin A, Shultz S, Jones W. Social visual engagement in infants and toddlers with autism: early developmental transitions and a model of pathogenesis. Neurosci Biobehav Rev 2015;50:189-203. [Crossref] [PubMed]
- Jones W, Klin A. Attention to eyes is present but in decline in 2-6-month-old infants later diagnosed with autism. Nature 2013;504:427-31. [Crossref] [PubMed]
- Bosl W, Tierney A, Tager-Flusberg H, et al. EEG complexity as a biomarker for autism spectrum disorder risk. BMC Med 2011;9:18. [Crossref] [PubMed]
- Canfield MA, Langlois PH, Rutenberg GW, et al. The association between newborn screening analytes and childhood autism in a Texas Medicaid population, 2010-2012. Am J Med Genet B Neuropsychiatr Genet 2019;180:291-304. [Crossref] [PubMed]
- Patriquin MA, Hartwig EM, Friedman BH, et al. Autonomic response in autism spectrum disorder: Relationship to social and cognitive functioning. Biol Psychol 2019;145:185-97. [Crossref] [PubMed]
- Klusek J, Roberts JE, Losh M. Cardiac autonomic regulation in autism and Fragile X syndrome: a review. Psychol Bull 2015;141:141-75. [Crossref] [PubMed]
- Bujnakova I, Ondrejka I, Mestanik M, et al. Autism spectrum disorder is associated with autonomic underarousal. Physiol Res 2016;65:S673-82. [PubMed]
- Nystrom P, Gliga T, Nilsson Jobs E, et al. Enhanced pupillary light reflex in infancy is associated with autism diagnosis in toddlerhood. Nat Commun 2018;9:1678. [Crossref] [PubMed]
- Frye RE, James SJ. Metabolic pathology of autism in relation to redox metabolism. Biomark Med 2014;8:321-30. [Crossref] [PubMed]
- Howsmon DP, Kruger U, Melnyk S, et al. Classification and adaptive behavior prediction of children with autism spectrum disorder based upon multivariate data analysis of markers of oxidative stress and DNA methylation. PLoS Comput Biol 2017;13:e1005385. [Crossref] [PubMed]
- Howsmon DP, Vargason T, Rubin RA, et al. Multivariate techniques enable a biochemical classification of children with autism spectrum disorder versus typically-developing peers: A comparison and validation study. Bioeng Transl Med 2018;3:156-65. [Crossref] [PubMed]
- Barone R, Alaimo S, Messina M, et al. A Subset of Patients With Autism Spectrum Disorders Show a Distinctive Metabolic Profile by Dried Blood Spot Analyses. Front Psychiatry 2018;9:636. [Crossref] [PubMed]
- Corbett BA, Kantor AB, Schulman H, et al. A proteomic study of serum from children with autism showing differential expression of apolipoproteins and complement proteins. Mol Psychiatry 2007;12:292-306. [Crossref] [PubMed]
- Momeni N, Bergquist J, Brudin L, et al. A novel blood-based biomarker for detection of autism spectrum disorders. Transl Psychiatry 2012;2:e91. [Crossref] [PubMed]
- Ngounou Wetie AG, Wormwood K, Thome J, et al. A pilot proteomic study of protein markers in autism spectrum disorder. Electrophoresis 2014;35:2046-54. [Crossref] [PubMed]
- Schwarz E, Guest PC, Rahmoune H, et al. Sex-specific serum biomarker patterns in adults with Asperger's syndrome. Mol Psychiatry 2011;16:1213-20. [Crossref] [PubMed]
- Shen L, Zhang K, Feng C, et al. iTRAQ-Based Proteomic Analysis Reveals Protein Profile in Plasma from Children with Autism. Proteomics Clin Appl 2018;12:e1700085. [Crossref] [PubMed]
- Skafidas E, Testa R, Zantomio D, et al. Predicting the diagnosis of autism spectrum disorder using gene pathway analysis. Mol Psychiatry 2014;19:504-10. [Crossref] [PubMed]
- Hicks SD, Rajan AT, Wagner KE, et al. Validation of a Salivary RNA Test for Childhood Autism Spectrum Disorder. Front Genet 2018;9:534. [Crossref] [PubMed]
- McPartland JC. Developing Clinically Practicable Biomarkers for Autism Spectrum Disorder. J Autism Dev Disord 2017;47:2935-7. [Crossref] [PubMed]
- Kang E, Keifer CM, Levy EJ, et al. Atypicality of the N170 Event-Related Potential in Autism Spectrum Disorder: A Meta-analysis. Biol Psychiatry Cogn Neurosci Neuroimaging 2018;3:657-66. [Crossref] [PubMed]
- McPartland JC, Wu J, Bailey CA, et al. Atypical neural specialization for social percepts in autism spectrum disorder. Soc Neurosci 2011;6:436-51. [Crossref] [PubMed]
- Tye C, Battaglia M, Bertoletti E, et al. Altered neurophysiological responses to emotional faces discriminate children with ASD, ADHD and ASD+ADHD. Biol Psychol 2014;103:125-34. [Crossref] [PubMed]
- McPartland J, Dawson G, Webb SJ, et al. Event-related brain potentials reveal anomalies in temporal processing of faces in autism spectrum disorder. J Child Psychol Psychiatry 2004;45:1235-45. [Crossref] [PubMed]
- O'Connor K, Hamm JP, Kirk IJ. The neurophysiological correlates of face processing in adults and children with Asperger's syndrome. Brain Cogn 2005;59:82-95. [Crossref] [PubMed]
- Bernier R, Dawson G, Webb S, et al. EEG mu rhythm and imitation impairments in individuals with autism spectrum disorder. Brain Cogn 2007;64:228-37. [Crossref] [PubMed]
- Oberman LM, Hubbard EM, McCleery JP, et al. EEG evidence for mirror neuron dysfunction in autism spectrum disorders. Brain Res Cogn Brain Res 2005;24:190-8. [Crossref] [PubMed]
- Martineau J, Cochin S, Magne R, et al. Impaired cortical activation in autistic children: is the mirror neuron system involved? Int J Psychophysiol 2008;68:35-40. [Crossref] [PubMed]
- Milne E, Scope A, Pascalis O, et al. Independent component analysis reveals atypical electroencephalographic activity during visual perception in individuals with autism. Biol Psychiatry 2009;65:22-30. [Crossref] [PubMed]
- Snijders TM, Milivojevic B, Kemner C. Atypical excitation-inhibition balance in autism captured by the gamma response to contextual modulation. Neuroimage Clin 2013;3:65-72. [Crossref] [PubMed]
- Brown C, Gruber T, Boucher J, et al. Gamma abnormalities during perception of illusory figures in autism. Cortex 2005;41:364-76. [Crossref] [PubMed]
- Stroganova TA, Orekhova EV, Prokofyev AO, et al. High-frequency oscillatory response to illusory contour in typically developing boys and boys with autism spectrum disorders. Cortex 2012;48:701-17. [Crossref] [PubMed]
- Sun L, Grutzner C, Bolte S, et al. Impaired gamma-band activity during perceptual organization in adults with autism spectrum disorders: evidence for dysfunctional network activity in frontal-posterior cortices. J Neurosci 2012;32:9563-73. [Crossref] [PubMed]
- Matsuzaki J, Ku M, Berman JI, et al. Abnormal auditory mismatch fields in adults with autism spectrum disorder. Neurosci Lett 2019;698:140-5. [Crossref] [PubMed]
- Port RG, Edgar JC, Ku M, et al. Maturation of auditory neural processes in autism spectrum disorder - A longitudinal MEG study. Neuroimage Clin 2016;11:566-77. [Crossref] [PubMed]
- Berman JI, Edgar JC, Blaskey L, et al. Multimodal Diffusion-MRI and MEG Assessment of Auditory and Language System Development in Autism Spectrum Disorder. Front Neuroanat 2016;10:30. [Crossref] [PubMed]
- Edgar JC, Khan SY, Blaskey L, et al. Neuromagnetic oscillations predict evoked-response latency delays and core language deficits in autism spectrum disorders. J Autism Dev Disord 2015;45:395-405. [Crossref] [PubMed]
- Shen MD, Nordahl CW, Li DD, et al. Extra-axial cerebrospinal fluid in high-risk and normal-risk children with autism aged 2-4 years: a case-control study. Lancet Psychiatry 2018;5:895-904. [Crossref] [PubMed]
- Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet 2008;9:341-55. [Crossref] [PubMed]
- Frye RE, Rossignol DA. Identification and Treatment of Pathophysiological Comorbidities of Autism Spectrum Disorder to Achieve Optimal Outcomes. Clin Med Insights Pediatr 2016;10:43-56. [Crossref] [PubMed]
- Rossignol DA, Frye RE. A review of research trends in physiological abnormalities in autism spectrum disorders: immune dysregulation, inflammation, oxidative stress, mitochondrial dysfunction and environmental toxicant exposures. Mol Psychiatry 2012;17:389-401. [Crossref] [PubMed]
- Schaefer GB, Mendelsohn NJ, Professional P, et al. Clinical genetics evaluation in identifying the etiology of autism spectrum disorders: 2013 guideline revisions. Genet Med 2013;15:399-407. [Crossref] [PubMed]
- Tammimies K, Marshall CR, Walker S, et al. Molecular Diagnostic Yield of Chromosomal Microarray Analysis and Whole-Exome Sequencing in Children With Autism Spectrum Disorder. JAMA 2015;314:895-903. [Crossref] [PubMed]
- Reddy KS. Cytogenetic abnormalities and fragile-X syndrome in Autism Spectrum Disorder. BMC Med Genet 2005;6:3. [Crossref] [PubMed]
- Weidmer-Mikhail E, Sheldon S, Ghaziuddin M. Chromosomes in autism and related pervasive developmental disorders: a cytogenetic study. J Intellect Disabil Res 1998;42:8-12. [Crossref] [PubMed]
- Vorstman JAS, Parr JR, Moreno-De-Luca D, et al. Autism genetics: opportunities and challenges for clinical translation. Nat Rev Genet 2017;18:362-76. [Crossref] [PubMed]
- Oikonomakis V, Kosma K, Mitrakos A, et al. Recurrent copy number variations as risk factors for autism spectrum disorders: analysis of the clinical implications. Clin Genet 2016;89:708-18. [Crossref] [PubMed]
- Rossi M, El-Khechen D, Black MH, et al. Outcomes of Diagnostic Exome Sequencing in Patients With Diagnosed or Suspected Autism Spectrum Disorders. Pediatr Neurol 2017;70:34-43.e2. [Crossref] [PubMed]
- Roesser J. Diagnostic yield of genetic testing in children diagnosed with autism spectrum disorders at a regional referral center. Clin Pediatr (Phila) 2011;50:834-43. [Crossref] [PubMed]
- Pain O, Pocklington AJ, Holmans PA, et al. Novel Insight Into the Etiology of Autism Spectrum Disorder Gained by Integrating Expression Data With Genome-wide Association Statistics. Biol Psychiatry 2019;86:265-73. [Crossref] [PubMed]
- Amaral DG, Li D, Libero L, et al. In pursuit of neurophenotypes: The consequences of having autism and a big brain. Autism Res 2017;10:711-22. [Crossref] [PubMed]
- Smith AM, King JJ, West PR, et al. Amino Acid Dysregulation Metabotypes: Potential Biomarkers for Diagnosis and Individualized Treatment for Subtypes of Autism Spectrum Disorder. Biol Psychiatry 2019;85:345-54. [Crossref] [PubMed]
- Novarino G, El-Fishawy P, Kayserili H, et al. Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science 2012;338:394-7. [Crossref] [PubMed]
- James SJ, Cutler P, Melnyk S, et al. Metabolic biomarkers of increased oxidative stress and impaired methylation capacity in children with autism. Am J Clin Nutr 2004;80:1611-7. [Crossref] [PubMed]
- James SJ, Melnyk S, Fuchs G, et al. Efficacy of methylcobalamin and folinic acid treatment on glutathione redox status in children with autism. Am J Clin Nutr 2009;89:425-30. [Crossref] [PubMed]
- Rose S, Melnyk S, Trusty TA, et al. Intracellular and extracellular redox status and free radical generation in primary immune cells from children with autism. Autism Res Treat 2012;2012:986519. [PubMed]
- James SJ, Rose S, Melnyk S, et al. Cellular and mitochondrial glutathione redox imbalance in lymphoblastoid cells derived from children with autism. FASEB J 2009;23:2374-83. [Crossref] [PubMed]
- Rose S, Melnyk S, Pavliv O, et al. Evidence of oxidative damage and inflammation associated with low glutathione redox status in the autism brain. Transl Psychiatry 2012;2:e134. [Crossref] [PubMed]
- Frustaci A, Neri M, Cesario A, et al. Oxidative stress-related biomarkers in autism: systematic review and meta-analyses. Free Radic Biol Med 2012;52:2128-41. [Crossref] [PubMed]
- Rossignol DA, Frye RE. Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol Psychiatry 2012;17:290-314. [Crossref] [PubMed]
- Rose S, Niyazov DM, Rossignol DA, et al. Clinical and Molecular Characteristics of Mitochondrial Dysfunction in Autism Spectrum Disorder. Mol Diagn Ther 2018;22:571-93. [Crossref] [PubMed]
- Goldenthal MJ, Damle S, Sheth S, et al. Mitochondrial enzyme dysfunction in autism spectrum disorders; a novel biomarker revealed from buccal swab analysis. Biomark Med 2015;9:957-65. [Crossref] [PubMed]
- Delhey L, Kilinc EN, Yin L, et al. Bioenergetic variation is related to autism symptomatology. Metab Brain Dis 2017;32:2021-31. [Crossref] [PubMed]
- Frye RE. Biomarkers of Abnormal Energy Metabolism in Children with Autism Spectrum Disorder. North American Journal of Medicine and Science 2012;5:141-7. [Crossref]
- Frye RE, Melnyk S, Macfabe DF. Unique acyl-carnitine profiles are potential biomarkers for acquired mitochondrial disease in autism spectrum disorder. Transl Psychiatry 2013;3:e220. [Crossref] [PubMed]
- Khemakhem AM, Frye RE, El-Ansary A, et al. Novel biomarkers of metabolic dysfunction is autism spectrum disorder: potential for biological diagnostic markers. Metab Brain Dis 2017;32:1983-97. [Crossref] [PubMed]
- El-Ansary A, Al-Daihan S, Al-Dabas A, et al. Activities of key glycolytic enzymes in the plasma of Saudi autistic patients. Open Access J Clin Trials 2010;10:49-57. [Crossref]
- El-Ansary A, Bjorklund G, Chirumbolo S, et al. Predictive value of selected biomarkers related to metabolism and oxidative stress in children with autism spectrum disorder. Metab Brain Dis 2017;32:1209-21. [Crossref] [PubMed]
- Al-Mosalem OA, El-Ansary A, Attas O, et al. Metabolic biomarkers related to energy metabolism in Saudi autistic children. Clin Biochem 2009;42:949-57. [Crossref] [PubMed]
- El-Ansary AK, Ben Bacha AG, Al-Ayadhi LY. Proinflammatory and proapoptotic markers in relation to mono and di-cations in plasma of autistic patients from Saudi Arabia. J Neuroinflammation 2011;8:142. [Crossref] [PubMed]
- Filipek PA, Juranek J, Nguyen MT, et al. Relative carnitine deficiency in autism. J Autism Dev Disord 2004;34:615-23. [Crossref] [PubMed]
- Mostafa GA, Al-Ayadhi LY. Reduced levels of plasma polyunsaturated fatty acids and serum carnitine in autistic children: relation to gastrointestinal manifestations. Behav Brain Funct 2015;11:4. [Crossref] [PubMed]
- Celestino-Soper PB, Violante S, Crawford EL, et al. A common X-linked inborn error of carnitine biosynthesis may be a risk factor for nondysmorphic autism. Proc Natl Acad Sci U S A 2012;109:7974-81. [Crossref] [PubMed]
- Guevara-Campos J, González-Guevara L, Guevara-González J, et al. First Case Report of Primary Carnitine Deficiency Manifested as Intellectual Disability and Autism Spectrum Disorder. Brain Sci 2019. [Crossref] [PubMed]
- Geier DA, Kern JK, Davis G, et al. A prospective double-blind, randomized clinical trial of levocarnitine to treat autism spectrum disorders. Med Sci Monit 2011;17:PI15-23. [Crossref] [PubMed]
- Fahmy SF, El-hamamsy MH, Zaki OK, et al. l-Carnitine supplementation improves the behavioral symptoms in autistic children. Research in Autism Spectrum Disorders 2013;7:159-66. [Crossref]
- Goin-Kochel RP, Scaglia F, Schaaf CP, et al. Side Effects and Behavioral Outcomes Following High-Dose Carnitine Supplementation Among Young Males With Autism Spectrum Disorder: A Pilot Study. Glob Pediatr Health 2019;6:2333794X19830696.
- Frye RE, Sequeira JM, Quadros EV, et al. Cerebral folate receptor autoantibodies in autism spectrum disorder. Mol Psychiatry 2013;18:369-81. [Crossref] [PubMed]
- Frye RE, Slattery J, Delhey L, et al. Folinic acid improves verbal communication in children with autism and language impairment: a randomized double-blind placebo-controlled trial. Mol Psychiatry 2018;23:247-56. [Crossref] [PubMed]
- de Magistris L, Familiari V, Pascotto A, et al. Alterations of the intestinal barrier in patients with autism spectrum disorders and in their first-degree relatives. J Pediatr Gastroenterol Nutr 2010;51:418-24. [Crossref] [PubMed]
- Fiorentino M, Sapone A, Senger S, et al. Blood-brain barrier and intestinal epithelial barrier alterations in autism spectrum disorders. Mol Autism 2016;7:49. [Crossref] [PubMed]
- Esnafoglu E, Cirrik S, Ayyildiz SN, et al. Increased Serum Zonulin Levels as an Intestinal Permeability Marker in Autistic Subjects. J Pediatr 2017;188:240-4. [Crossref] [PubMed]
- Dalton N, Chandler S, Turner C, et al. Gut permeability in autism spectrum disorders. Autism Res 2014;7:305-13. [Crossref] [PubMed]
- Lynch GTF, James SM, VanDam M. Pupillary Response and Phenotype in ASD: Latency to Constriction Discriminates ASD from Typically Developing Adolescents. Autism Res 2018;11:364-75. [Crossref] [PubMed]
- Kuiper MWM, Verhoeven EWM, Geurts HM. Heart rate variability predicts inhibitory control in adults with autism spectrum disorders. Biol Psychol 2017;128:141-52. [Crossref] [PubMed]
- Thapa R, Alvares GA, Zaidi TA, et al. Reduced heart rate variability in adults with autism spectrum disorder. Autism Res 2019;12:922-30. [Crossref] [PubMed]
- Dijkhuis RR, Ziermans T, van Rijn S, et al. Emotional Arousal During Social Stress in Young Adults With Autism: Insights From Heart Rate, Heart Rate Variability and Self-Report. J Autism Dev Disord 2019;49:2524-35. [Crossref] [PubMed]
- Cai RY, Richdale AL, Dissanayake C, et al. Resting heart rate variability, emotion regulation, psychological wellbeing and autism symptomatology in adults with and without autism. Int J Psychophysiol 2019;137:54-62. [Crossref] [PubMed]
- Harder R, Malow BA, Goodpaster RL, et al. Heart rate variability during sleep in children with autism spectrum disorder. Clin Auton Res 2016;26:423-32. [Crossref] [PubMed]
- Ferguson BJ, Marler S, Altstein LL, et al. Psychophysiological Associations with Gastrointestinal Symptomatology in Autism Spectrum Disorder. Autism Res 2017;10:276-88. [Crossref] [PubMed]
- Wang Y, Hensley MK, Tasman A, et al. Heart Rate Variability and Skin Conductance During Repetitive TMS Course in Children with Autism. Appl Psychophysiol Biofeedback 2016;41:47-60. [Crossref] [PubMed]
- Ruiz-Robledillo N, Bellosta-Batalla M, Moya-Albiol L. Lower cardiovascular reactivity to acute stress in informal caregivers of people with autism spectrum disorder than in non-caregivers: Implications for health outcomes. Int J Psychophysiol 2015;98:143-50. [Crossref] [PubMed]
- Factor RS, Swain DM, Scarpa A. Child Autism Spectrum Disorder Traits and Parenting Stress: The Utility of Using a Physiological Measure of Parental Stress. J Autism Dev Disord 2018;48:1081-91. [Crossref] [PubMed]
- Pattini E, Carnevali L, Troisi A, et al. Psychological characteristics and physiological reactivity to acute stress in mothers of children with autism spectrum disorder. Stress Health 2019. [Crossref] [PubMed]
- Vidyashree HM, Maheshkumar K, Sundareswaran L, et al. Effect of Yoga Intervention on Short-Term Heart Rate Variability in Children with Autism Spectrum Disorder. Int J Yoga 2019;12:73-7. [Crossref] [PubMed]
- Casanova MF, Hensley MK, Sokhadze EM, et al. Effects of weekly low-frequency rTMS on autonomic measures in children with autism spectrum disorder. Front Hum Neurosci 2014;8:851. [Crossref] [PubMed]
- Dinalankara DMR, Miles JH, Nicole Takahashi T, et al. Atypical pupillary light reflex in 2-6-year-old children with autism spectrum disorders. Autism Res 2017;10:829-38. [Crossref] [PubMed]
- Daluwatte C, Miles JH, Sun J, et al. Association between pupillary light reflex and sensory behaviors in children with autism spectrum disorders. Res Dev Disabil 2015;37:209-15. [Crossref] [PubMed]
- Daluwatte C, Miles JH, Christ SE, et al. Atypical pupillary light reflex and heart rate variability in children with autism spectrum disorder. J Autism Dev Disord 2013;43:1910-25. [Crossref] [PubMed]
- Fan X, Miles JH, Takahashi N, et al. Abnormal transient pupillary light reflex in individuals with autism spectrum disorders. J Autism Dev Disord 2009;39:1499-508. [Crossref] [PubMed]
- Benevides TW, Lane SJ. A review of cardiac autonomic measures: considerations for examination of physiological response in children with autism spectrum disorder. J Autism Dev Disord 2015;45:560-75. [Crossref] [PubMed]
- Quintana DS, Alvares GA, Heathers JA. Guidelines for Reporting Articles on Psychiatry and Heart rate variability (GRAPH): recommendations to advance research communication. Transl Psychiatry 2016;6:e803. [Crossref] [PubMed]
- Frye RE, Melnyk S, Fuchs G, et al. Effectiveness of methylcobalamin and folinic Acid treatment on adaptive behavior in children with autistic disorder is related to glutathione redox status. Autism Res Treat 2013;2013:609705. [PubMed]
- Hendren RL, James SJ, Widjaja F, et al. Randomized, Placebo-Controlled Trial of Methyl B12 for Children with Autism. J Child Adolesc Psychopharmacol 2016;26:774-83. [Crossref] [PubMed]
- Delhey LM, Tippett M, Rose S, et al. Comparison of Treatment for Metabolic Disorders Associated with Autism:Reanalysis of Three Clinical Trials. Front Neurosci 2018;12:19. [Crossref] [PubMed]
- Frye RE, DeLatorre R, Taylor HB, et al. Metabolic effects of sapropterin treatment in autism spectrum disorder: a preliminary study. Transl Psychiatry 2013;3:e237. [Crossref] [PubMed]
- Connery K, Tippett M, Delhey LM, et al. Intravenous immunoglobulin for the treatment of autoimmune encephalopathy in children with autism. Transl Psychiatry 2018;8:148. [Crossref] [PubMed]
- Frye RE. Social Skills Deficits in Autism Spectrum Disorder: Potential Biological Origins and Progress in Developing Therapeutic Agents. CNS Drugs 2018;32:713-34. [Crossref] [PubMed]
- Zamzow RM, Ferguson BJ, Ragsdale AS, et al. Effects of acute beta-adrenergic antagonism on verbal problem solving in autism spectrum disorder and exploration of treatment response markers. J Clin Exp Neuropsychol 2017;39:596-606. [Crossref] [PubMed]
- Sauer C, Montag C, Worner C, et al. Effects of a common variant in the CD38 gene on social processing in an oxytocin challenge study: possible links to autism. Neuropsychopharmacology 2012;37:1474-82. [Crossref] [PubMed]
- Kosaka H, Okamoto Y, Munesue T, et al. Oxytocin efficacy is modulated by dosage and oxytocin receptor genotype in young adults with high-functioning autism: a 24-week randomized clinical trial. Transl Psychiatry 2016;6:e872. [Crossref] [PubMed]
- Ajram LA, Horder J, Mendez MA, et al. Shifting brain inhibitory balance and connectivity of the prefrontal cortex of adults with autism spectrum disorder. Transl Psychiatry 2017;7:e1137. [Crossref] [PubMed]
- Fox-Edmiston E, Van de Water J. Maternal Anti-Fetal Brain IgG Autoantibodies and Autism Spectrum Disorder: Current Knowledge and its Implications for Potential Therapeutics. CNS Drugs 2015;29:715-24. [Crossref] [PubMed]
- Hallmayer J, Cleveland S, Torres A, et al. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry 2011;68:1095-102. [Crossref] [PubMed]
- Beversdorf DQ, Missouri Autism Summit C. Phenotyping, Etiological Factors, and Biomarkers: Toward Precision Medicine in Autism Spectrum Disorders. J Dev Behav Pediatr 2016;37:659-73. [Crossref] [PubMed]
- Buie T, Campbell DB, Fuchs GJ 3rd, et al. Evaluation, diagnosis, and treatment of gastrointestinal disorders in individuals with ASDs: a consensus report. Pediatrics 2010;125 Suppl 1:S1-18. [Crossref] [PubMed]
- Kozak MJ, Cuthbert BN. The NIMH Research Domain Criteria Initiative: Background, Issues, and Pragmatics. Psychophysiology 2016;53:286-97. [Crossref] [PubMed]


