
Genome-wide profiling of prognosis-related alternative splicing signatures in sarcoma
Introduction
Alternative splicing (AS) is a ubiquitous regulatory mechanism that enables eukaryotic cells to generate vast and diverse mature mRNAs from a limited number of genes. AS remarkably increases both transcriptome and proteome complexity via different precursor mRNA splicing processes (1,2). Throughout the whole developmental stage, AS plays a crucial role in the regulation of cell differentiation, cell-specific functions, and cell senescence to further control growth and metabolism (3-7). Given this, multiple pathological processes, including cancers, are also produced because of AS deregulation. Aberrant AS may initiate loss-of-function in tumor suppressors, the activation of oncogenes or cancer pathways to affect tumorigenesis, development, and prognosis through abnormal proliferation, altered epigenetics, and resistance to apoptosis and chemotherapy (8-12). Therefore, AS events have been proposed as ideal biomarkers for cancer diagnosis and prognosis and even potential targets for new anti-carcinogen discoveries (8,10,12,13).
Sarcomas (SARCs), which arise from mesenchymal tissues, are malignant tumors comprising less than 10% of all cancers (14). Statistically, approximately 10,700 diagnoses and 3,800 deaths per year in the US are relevant to SARC, while in Europe, SARC represents only 1–2% of all cancers in adults with an overall annual incidence of 5.6 cases per 100,000 adults (15,16). Compared to the low incidence in adults, SARCs account for a higher percentage of overall cancer morbidity and mortality in children and young adults (ages 20–39). SARCs are a group of heterogeneous mesenchymal malignancies with over eighty histological subtypes, which are broadly categorized as soft-tissue SARCs and bone SARCs (17). Clinically, surgical resection, radiotherapy, and chemotherapy adopted singly or together be effective in treating specific localized SARCs. However, for many SARC patients who have chemotherapy resistance or metastatic foci, new therapeutic regimens are needed.
Along with the discovery of an increasing number of pivotal genes and biological mechanisms underlying carcinogenesis and disease progression, correlative pharmacologic and genetic therapies based on these are becoming progressively essential in routine treatment. In turn, molecular characteristics gradually guide the nosological classification of SARCs. Hence, it is imperative to explore molecular mechanisms more deeply in the prognosis of SARCs. Several studies have been undertaken to expand the genomic diversity of oncogenic drivers and identify potential therapeutic targets in SARC through genome-scale analyses of mRNAs, microRNAs, and protein, and alterations in DNA sequences, methylation, and copy numbers (18,19). AS is worth further exploring as a new field, as it is an indispensable part in the molecular mechanisms of SARCs.
In recent years, many studies of AS have mainly focused on distinguishing “SARC-specific” AS events by molecular experiments (20-23). However, comprehensive analyses of survival-related AS events are still scarce in SARC. Powered by high-throughput RNA-seq, the investigation of cancer-related AS events and counterpart molecular mechanisms in a relatively larger population has become obtainable with the rapid accumulation of transcriptome data and clinical data (24,25). Through the analysis of genome-wide profiling, prognostic AS signatures have been reported in many cancers, such as non-small cell lung cancer, colorectal cancer, prostate adenocarcinoma, esophageal carcinoma, and gastrointestinal pan-adenocarcinomas (26-31).
The Cancer Genome Atlas (TCGA) project provides a rich source for the research of AS in cancer with mass data of transcript isoforms, corresponding genes, and the survival status of tumor patients (24). We systematically analyzed genome-wide AS events in the SARC cohort from TCGA using a series of bioinformatics methods to obtain a certain amount of survival-related AS events and revealed the latent splicing networks. More importantly, high-performance prognostic predictors were constructed to evaluate the potential of AS events in the prognosis prediction of SARC. These findings, for the first time, supply systematic novel insights into the latent functions of SARC-specific AS events.
Methods
AS event curation from TCGA RNA-seq data
RNA-seq data from the TCGA SARC cohorts were available from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/).
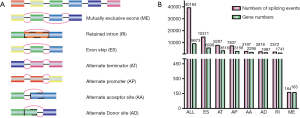
TCGASpliceSeq (http://bioinformatics.mdanderson.org/TCGASpliceSeq) is a web-based resource that includes the mRNA AS patterns of tumors from TCGA (32). We used this tool to analyze the different AS patterns of TCGA SARC samples. The SpliceSeq program aligns to the gene’s protein-coding transcripts in the ensemble gene database and conveniently generates a unified splice graph. Then, the splice graph is aligned to the TCGA sample reads, and summary statistics are collected for each transcript isoform. SpliceSeq can calculate the percent-spliced-in (PSI) value for each AS event, which supplies a clear view of splice junctions and the proportion of exons. The PSI value is a standard, intuitive ratio for quantifying seven types of AS events (33): exon skip (ES), retained intron (RI), mutually exclusive exons (MEs), alternate promoter (AP), alternate terminator (AT), alternate donor site (AD), and alternate acceptor site (AA). Only AS events with a PSI value >75% and a standard deviation >0.1 were included in the present analysis. A schematic diagram illustrating the seven types of AS events are shown in Figure 1A.
Survival analysis
A total of 261 SARC patients with detailed overall survival (OS) information were included in this study. The summary characteristics of these patients are depicted in Table S1. Univariate Cox regression analysis was performed to identify survival-related AS events in SARC with P values less than 0.05. Then, multivariate Cox regression analysis was further employed to determine whether splicing events were independent prognostic factors. Finally, the risk score for the seven types of AS events was calculated for survival prediction. Furthermore, predictive models of Kaplan-Meier (K-M) survival analysis and receiver operating characteristic (ROC) curves were constructed to evaluate the clinical value of the risk scores. The area under the ROC curve (AUC) was used to assess the predictive accuracy of the prognostic models.
Full table
UpSet plot and gene interaction network
The intersections between the different types of AS events were also investigated by the UpSet package of R software. UpSet plots are a more intuitive and superior alternative to Venn diagrams for visualizing intersecting sets (34). After selection by univariate Cox regression analysis, the candidate genes of prognosis-related AS events with P value <0.005 were submitted to Metascape (http://metascape.org) for protein-protein interaction (PPI) analysis (35,36). The molecular complex detection (MCODE) algorithm was then applied to identify neighborhoods where proteins were densely connected in the PPI network. Subsequently, Gene Ontology (GO) enrichment analysis was performed for each MCODE network to assign biological interpretations to the network components via Metascape. We further set the P value to less than 0.05 in the univariate Cox regression analysis to obtain genes of survival-related AS events for the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and GO analyses performed by Metascape. The ggplot2 package of R software was used to visualize the KEGG pathway enrichment analysis.
Splicing correlation network construction
The association between survival-related AS events and splicing factors (SFs) was further investigated. A list of known SFs was extracted from the SpliceAid 2 (www.introni.it/spliceaid.html) database (37). The level 3 expression profiles of SFs were downloaded from the TCGA database and converted into transcripts per million (TPM), which is considered a more reasonable data format for RNA-seq (38). After that, the log-rank test was conducted to select survival-related SFs. Pearson’s correlation test was further applied to assess the association between survival-related AS events and SFs.
DriverDBv2 (a database for human cancer driver gene research; http://driverdb.tms.cmu.edu.tw/driverdbv2/) was used to discover driver genes and their mutational types (39). Pearson’s correlation test was performed to investigate the correlation between the expression of driver genes and AS events. Also, t-test was conducted to find the statistical significance between gene mutation and AS events. All statistical analyses were performed using R/Bioconductor (version 3.4.3) and reported P values of <0.05 were considered statistically significant (P values were two-sided).
Results
Integrated mRNA splicing event profiles in the SARC cohort of TCGA
All splicing events were calculated by SpliceSeq. A total of 9,673 genes with 40,184 AS events were detected in 261 sarcomatous samples, including 15,311 ESs in 6,038 genes, 8,287 ATs in 3,616 genes, 7,837 APs in 3,156 genes, 3,197 AAs in 2,295 genes, 2,816 ADs in 1,987 genes, 2,572 RIs in 1,741 genes and 164 MEs in 163 genes (Figure 1B). Overall, the results showed that one gene might have an average of 4.1 AS events. Among these splicing subtypes, ES was the main subtype of AS events, while MEs were rare in sarcomas. Also, only a small fraction of all AS events (1,438 out of 40,184) were novel splices. The summary clinical characteristics of the SARC patients are shown in Table 1.

Full table
Survival-associated AS events in the SARC cohort
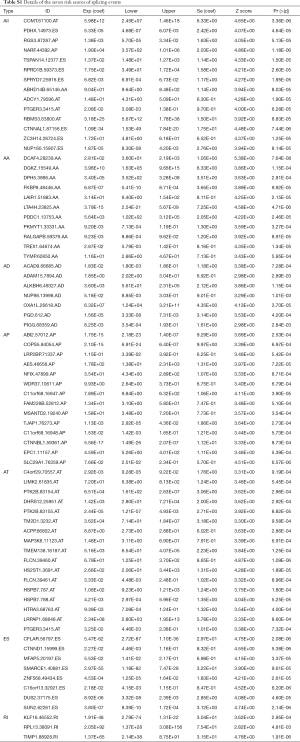
Univariate Cox analyses of OS were applied to find survival-associated AS events in the SARC cohort. The results showed that a total of 3,610 AS events from 2,291 genes were significantly associated with patient OS (P<0.05), including 203 RIs from 185 genes, 252 AAs from 240 genes, 252 Ads from 229 genes, 758 APs from 444 genes, 776 ATs from 423 genes, 1,359 ESs from 1,083 genes, and 10 MEs from 10 genes (Figure 2A). As depicted in the UpSet plot, most genes had only one prognosis-related AS subtype, while some genes had two or more AS subtypes associated with survival. Among these survival-related genes, 293 genes had more than one type of AS event, but none of them had the seven AS subtypes simultaneously (Figure 2B).
Gene network construction and functional enrichment analysis
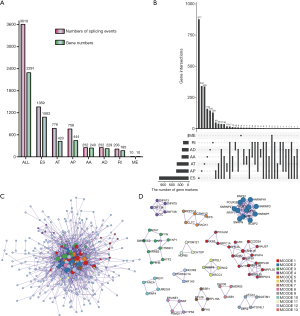
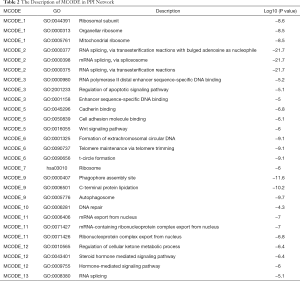
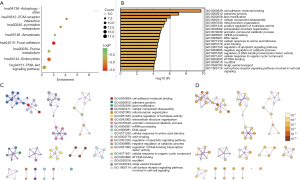
To reveal the interaction effects among the genes of prognosis-related AS events, a PPI network was constructed by Metascape (Figure 2C). After the molecular complex detection analysis, thirteen MCODE components identifying neighborhoods where proteins were densely connected were extracted from the PPI network (Figure 2D). Also, the GO enrichment analysis of each MCODE network is depicted in Table 2, in which the top three GO results are displayed. Among these thirteen MCODEs, the most significant part was MCODE2, which included twelve genes: RNPS1, POLR2E, SNRNP27, SRSF2, SNRPN, NCBP2, HNRNPF, HNRNPD, HNRNPH1, CPSF3, PPIE, and SRSF7. As shown in Table 2, these genes involved in MCODE2 were enriched in reactions with bulged adenosine as nucleophile, mRNA splicing via the spliceosome, and RNA splicing via transesterification reactions.
Full table
To analyze the biological classification of the survival-related AS genes, KEGG pathway and GO enrichment analyses were performed. The KEGG pathway analysis revealed that “focal adhesion” was the most significantly enriched by these genes, followed by “endocytosis” (Figure 3A). GO analysis revealed that “cell adhesion molecule binding,” “adherens junction” and “lipid modification” were the most significantly enriched (Figure 3B). As showed in the hierarchical cluster tree, the highest Kappa-statistical similarities existed in the “adherens junction” among gene memberships (Figure 3C), while the “cell adhesion molecule binding” was the most statistically significant (Figure 3D).
Construction of AS-based prognostic predictors
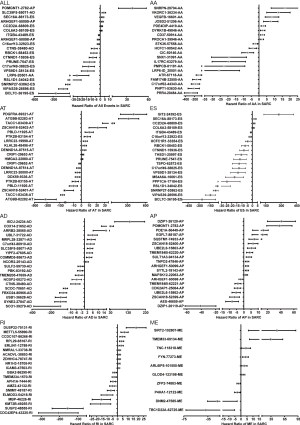
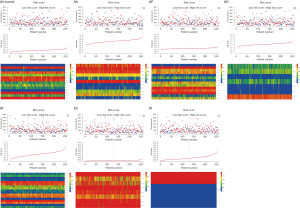
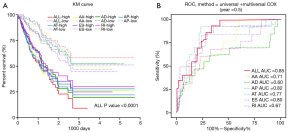
To clinically evaluate the prognostic value of AS in SARC, we constructed prognostic multivariate Cox regression models based on the top twenty significantly prognosis-related events in each AS subtype (Figure 4). The hazard ratio of each AS subtype was processed logarithmically to base 10. Owing to the quantitative differences in survival-associated AS events, we chose different p values to construct best risk scores with regard to each AS subtype, which included all splicing types (P<0.0001), AA (P<0.001), AP (P<0.001), ES (P<0.0001), RI (P<0.005), AD (P<0.005), and AT (P<0.001). However, the risk score of the ME type could not be constructed because AS events of the ME type were insufficient. The details of the AS events used for constructing the seven risk scores are shown in Table S1. SARC patients were classified into low-risk and high-risk groups according to the median value of the risk score as the cut-off (Figure 5). The Kaplan-Meier analysis proved that the survival time was diverse between the two groups in the Cox regression model of each AS subtype, excluding the ME type (P value <0.0001). Then, we integrated the six types of AS events and developed a comprehensive prognostic predictor, which also showed moderate performance in predicting prognosis (P value <0.0001) (Figure 6A). Notably, the ROC curve analysis confirmed that the integrated prognostic predictor displayed more of an ideal performance than the other models based on a single specific type of AS event. The AUC of the ROC curve for the integrated prognostic predictor was 0.85, followed by the AP and ES models with AUCs of 0.82 and 0.80, respectively (Figure 6B).
Survival-associated SFs and driver genes
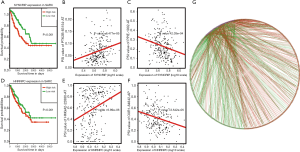
A total of 71 SFs from the SpliceAid 2 database were selected for survival analysis, and 26 survival-related SFs were identified (P<0.001). Furthermore, Spearman’s test was performed to explore the correlations between the PSI values of top significant AS events and prognosis-related SFs. Only relationships with the significant association (P<0.001) were included in the correlative network (Figure 7). As depicted in the network, most SFs were positively correlated with risk survival prognostic AS events while negatively correlated with protective survival prognostic AS events (Figure 7G). For example, the high expression of the SFs SYNCRIP and HNRNPC were significantly associated with poor survival prognosis (Figure 7A,D). In concordance, as shown in dot plots (Figure 7B,E), risk survival prognostic AS events such as the AT of PTK2B and AT of HMGA2 were positively correlated with the SFs SYNCRIP and HNRNPC, respectively. In contrast, protective survival prognostic AS events such as the AT of PPIE and AP of OXR1 were negatively correlated with the SFs SYNCRIP and HNRNPC (Figure 7C,F) respectively, which showed poor survival prognosis under high expression as well.
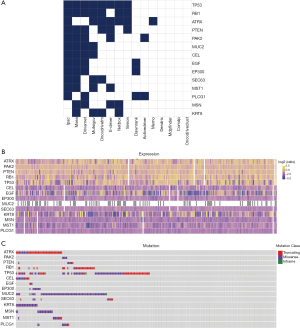
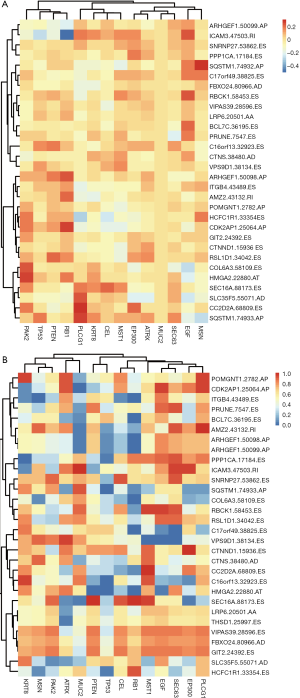
A list of driver genes was generated by at least four bioinformatics tools using the DriverDBv2, which is a database for cancer driver genes and mutations. The results showed that a total of 14 driver genes were identified (Figure S1A). In the mutation profile of the driver genes, mutations in TP53, RB1, and ATRX occurred in most of the SARC cohort from TCGA. Regarding mutation classification, truncating and missense mutations were the two main types for the driver genes TP53, RB1, ATRX, and MUC2 (Figure S1B,C). The correlations between the expression of the driver genes and PSI values of the top 30 survival-related AS events were evaluated. The results revealed that expression of PLCG1, PAK2, and RB1 was closely related to most of the top 30 survival-associated AS events with a high correlation coefficient (Figure S2A). Also, we conducted t-test to confirm the statistical significance of AS events and mutation profiles and found that the mutation status of TP53 and RB1 significantly correlated with most of the top 30 survival-associated AS events (Figure S2B).
Discussion
Considerable investigations have confirmed that by altering the composition of the proteome, the perturbation in mRNA AS patterns trigger the initiation and progression of diverse cancers (11,12). In the present study, we screened reasonable quantities of prognosis-related AS events and correlative SFs in SARC patients. Corresponding SF-AS event networks were also set up to elucidate potential regulatory mechanisms for the abnormalities in SARC. The genes of survival-related AS events were found to be enriched in several metabolic pathways and interacted closely with each other, especially in dense regions of the PPI network. We also implemented correlation analyses between variant driver genes and survival-associated AS events with the intent to guide further mutual mechanistic studies. Remarkably, all prognostic predictors we constructed based on the most significant prognosis-related AS events showed superior performance, especially the integrated prognostic predictor. Our comprehensive investigation first focused on the AS event characteristics at the genome-wide level to evaluate the potential of survival-related AS events as prognostic, predictive factors, and therapeutic target molecules in SARC.
A single type of pre-mRNA can be spliced in different ways to engender similar but not identical mature mRNA patterns. Nevertheless, the proteins translated by these homologous transcript isoforms may exert analogous or completely contrary functions in normal physiological conditions. For example, polypyrimidine tract binding protein 1 (PTBP1) is expressed at high levels during the mitotic period of neural stem cells, but upon neuronal differentiation, it becomes repressed and allows the induction of PTBP2 expression (40). Also, it has been demonstrated that the ubiquitous (Mef2Da1) and the muscle-specific (Mef2Da2) splicing isoforms antagonistically regulate muscle cell differentiation (41). Accumulating evidence has proven that aberrant splicing is often interrelated with the activation of oncogenes and the inactivation of tumor-suppressor genes. In one aspect, dysregulated splicing induces the activation of oncogenes by producing novel splice variants with tumorigenic properties. MDM2-ALT1, an isoform of the oncogene murine double minute 2 (MDM2), is typically induced in response to genotoxic stress. It is pervasive in various aggressive metastatic cancers, including pediatric rhabdomyosarcoma and lymphoma (42,43). In another aspect, the imbalanced components of splice variants from the same gene can affect the related downstream regulatory network, which also contributes to tumor development. For instance, EWS-FLI1, a fusion of Ewing’s sarcoma (EWS) and FLI1 genes, stimulates transcription of the proto-oncogene CCND1 encoding cyclin D1a and a less abundant but more oncogenic isoform, D1b.
In contrast to the EWS oncogene preferring the D1a isoform, EWS-FLI1 elevates the D1b/D1a transcript ratio, accelerating EWS cell growth (44). Vascular endothelial growth factor (VEGF), well known as a proangiogenic factor, can also confer antiangiogenic effects due to the divergent splice site choice. Its proangiogenic isoform (VEGF165) and antiangiogenic isoform (VEGF165b) are generated from proximal or distal splice site selections in exon 8, respectively. Any disturbance to the coordination of the pro- and antiangiogenic isoforms of VEGF can interfere with the maintenance of internal homeostasis, even resulting in angiogenic pathologies in different cancers (45). It is difficult to determine whether a gene is a tumor suppressor or oncogene since variant AS events from the same pre-mRNA may have similar or even opposite biological functions because of minute sequence differences. Therefore, multiple functions of a versatile gene should be discriminated depending on the specific predominant AS events.
In the present study, a series of survival-related AS events were identified via high-throughput technology. It was noted that most of these survival-associated AS events were adverse prognostic signatures in SARC. We used the most significant prognosis-related genes to construct a PPI network, from which thirteen MCODEs were highlighted, indicating potential molecular complexes. Thereinto, MCODE2 was the most statistically significant with GO enrichment in reactions with bulged adenosine as a nucleophile, mRNA splicing via the spliceosome and RNA splicing via transesterification reactions. With the assistance of the spliceosome, a multi-subunit RNA-protein complex comprising small nuclear RNAs (snRNAs) and spliceosomal protein, pre-mRNA splicing encompasses two sequential transesterification reactions (branching and exon ligation) to remove introns to produce mature mRNAs with uninterrupted protein-coding sequences (46,47). In the first transesterification step of splicing, the U2 snRNP interacts directly with the pre-mRNA branch point adenosine acting as the nucleophile within the bulged duplex formed between them (48,49). Based on the above physiological process, we have enough evidence to speculate that the survival-related genes of AS enriched in MCODE2 are linked to sarcomas through intricate splicing.
According to the results of the KEGG enrichment analysis, we found that focal adhesion was the most significant interference pathway related to AS. Multifunctional focal adhesion complexes facilitating contact between the extracellular matrix and actin cytoskeleton play an important role in controlling the cellular morphology and cytoplasmic signaling for survival, proliferation, differentiation, and motility. The uniqueness of focal adhesions rests with the cooperative connection between integrins and growth factor receptors accessing the cytoplasmic signaling network (50). As a key mediator for downstream signaling of integrins and growth factor receptors, focal adhesion kinase (FAK) has been indicated to be upregulated in many cancers (51,52). FAK plays a prominent role in tumor initiation, progression, and metastasis via its regulation of both tumor cells and their microenvironments, including tumor cell migration, invasion, angiogenesis and epithelial to mesenchymal transition (53). Moreover, it has been reported that FAK splice variants were experimentally detected in promoting cancer cell proliferation and migration in leukemia, non-small cell lung cancer and breast cancer (54-56).
Due to the high malignancy and mortality of SARCs, it is urgent to explore effective prognostic signatures and therapeutic targets in SARC more deeply. Some prognosis-related molecules for SARC have been consecutively proposed in recent years, such as mRNAs (57), miRNAs (58) and lncRNAs (59,60). With advances in high-throughput RNA-seq, several researchers have illuminated the prognostic value of AS events in multiple cancers at the genome-wide level (26-31). They suggested that AS events could be preeminent hallmarks for cancer prognosis prediction and further potential therapeutic targets. Hence, we equally used high-throughput RNA-seq to integrate clinical outcome data and AS events, aiming to offer a comprehensive recognition of prognosis-related AS events in SARC. According to the high accuracy of the prediction model we constructed, the signatures of survival associated AS events are ideal for predicting the prognosis of SARC.
As requisite regulatory molecules, SFs assist the spliceosome to successfully recognize and bind to specific pre-mRNA sequences, which subsequently promotes the generation of mature mRNA (61). Hence, it is imperative to uncover the regulatory network between AS events and SFs. In our research, after screening the most significant prognosis-related SFs and AS events, we constructed an SF-AS regulatory network. Interestingly, there was a general trend that most SFs were positively correlated with adverse AS events while negatively correlated with favorable AS events. Also, as observed in the intricate network, limited SFs dominated AS in a synergistic or antagonistic manner to engender an enormous amount of AS events. SFs comprise the serine/arginine-rich proteins (SR proteins) and the heterogeneous nuclear ribonucleoprotein (hnRNP) families. Physiologically, these two families perform opposite functions in splicing and bind to enhancer sites and silencer sites, respectively. It has been widely reported that dysregulated expression levels of SFs with or without mutations can affect cancer pathogenesis (13). However, the potential molecular interaction and detailed regulatory network of SFs during the AS process remain unclear, and more attention should be paid to future experimental verification.
SARCs are groups of histologically heterogeneous tumors that have historically complicated driver gene discovery. With a better understanding of the molecular mechanism of SARC, an increasing number of essential driver genes have been identified, including TP53, PIK3CA, SETD2, AKT1, FBXW7, FGFR1, FOXA2 and PDK1 (62-65). Subsequent signal pathway changes induced by mutations in such genes lead to accelerated cancer development and progression. Also, SF genes are significantly differentially expressed between cancer and corresponding normal samples and have reduced interindividual expression variation in cancer (66).
However, few studies have been conducted on the association between driver genes and AS events. Potential driver genes were screened by a bioinformatic tool in the present study, and we confirmed that mutations in TP53, RB1, and ATRX occurred in most of the SARC cohort from TCGA. Concordantly, these three driver genes were also included in fourteen genes obtained by the exome sequencing analysis of osteosarcoma (67) and are highly recurrently mutated across sarcoma types (18). Through the correlation analyses between the expression of driver genes and top survival-related AS events, RB1 was found to have a high correlation coefficient and a significant correlation simultaneously. Our study findings built a bridge between driver genes and AS events in SARC, and the mutually concrete regulatory mechanisms still need to be further expounded.
However, several limitations should be considered. First, the number of patients included in the SARC cohort was small. Second, the prognostic predictors proposed here have not been reproducibly verified due to the lack of another independent cohort of SARC patients. Third, further experimental validation should be performed in the future.
Conclusions
In summary, for the first time, our comprehensive research focused on extensive data on AS events in SARCs at the genome-wide level. The interpretation of aberrant AS patterns and corresponding regulatory networks may improve the management of SARCs, particularly broadening the novel field of prognosis prediction and underlying molecular-targeted therapies.
Acknowledgments
Funding: This grant of the study was from the National Natural Science Foundation of China (Grant No. 81572628) and the Guangdong Province Science and Technology plan project (Grant No. 2014A020212304). This funding made a significant contribution to study design, data interpretation, and writing.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work (including full data access, integrity of the data and the accuracy of the data analysis) in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This article does not contain any studies with human participants or animals performed by any of the authors. Therefore, informed consent is not needed.
References
- Yang X, Coulombe-Huntington J, Kang S, et al. Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing. Cell 2016;164:805-17. [Crossref] [PubMed]
- Liu Y, Gonzalez-Porta M, Santos S, et al. Impact of Alternative Splicing on the Human Proteome. Cell Rep 2017;20:1229-41. [Crossref] [PubMed]
- Porter RS, Jaamour F, Iwase S. Neuron-specific alternative splicing of transcriptional machineries: Implications for neurodevelopmental disorders. Molecular and Cellular Neuroscience 2018;87:35-45. [Crossref] [PubMed]
- Gallego-Paez LM, Bordone MC, Leote AC, et al. Alternative splicing: the pledge, the turn, and the prestige: The key role of alternative splicing in human biological systems. Hum Genet 2017;136:1015-42. [Crossref] [PubMed]
- Fiszbein A, Kornblihtt AR. Alternative splicing switches: Important players in cell differentiation. Bioessays 2017.39. [PubMed]
- Deschênes M, Chabot B. The emerging role of alternative splicing in senescence and aging. Aging Cell 2017;16:918-33. [Crossref] [PubMed]
- Baralle FE, Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol 2017;18:437-51. [Crossref] [PubMed]
- Song X, Zeng Z, Wei H, et al. Alternative splicing in cancers: From aberrant regulation to new therapeutics. Semin Cell Dev Biol 2018;75:13-22. [Crossref] [PubMed]
- Siegfried Z, Karni R. The role of alternative splicing in cancer drug resistance. Curr Opin Genet Dev 2018;48:16-21. [Crossref] [PubMed]
- Pradella D, Naro C, Sette C, et al. EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer 2017;16:8. [Crossref] [PubMed]
- Climente-González H, Porta-Pardo E, Godzik A, et al. The Functional Impact of Alternative Splicing in Cancer. Cell Rep 2017;20:2215-26. [Crossref] [PubMed]
- Chen J, Weiss WA. Alternative splicing in cancer: implications for biology and therapy. Oncogene 2015;34:1-14. [Crossref] [PubMed]
- Jyotsana N, Heuser M. Exploiting differential RNA splicing patterns: a potential new group of therapeutic targets in cancer. Expert Opin Ther Targets 2018;22:107-21. [Crossref] [PubMed]
- Helman LJ, Meltzer P. Mechanisms of sarcoma development. Nat Rev Cancer 2003;3:685-94. [Crossref] [PubMed]
- Stiller CA, Trama A, Serraino D, et al. Descriptive epidemiology of sarcomas in Europe: report from the RARECARE project. Eur J Cancer 2013;49:684-95. [Crossref] [PubMed]
- Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin 2009;59:225-49. [Crossref] [PubMed]
- Doyle LA. Sarcoma classification: an update based on the 2013 World Health Organization Classification of Tumors of Soft Tissue and Bone. Cancer 2014;120:1763-74. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas. Cell 2017;171:950-965.e28. [Crossref] [PubMed]
- Barretina J, Taylor BS, Banerji S, et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet 2010;42:715-21. [Crossref] [PubMed]
- Zhang M, Zhu B, Davie J. Alternative splicing of MEF2C pre-mRNA controls its activity in normal myogenesis and promotes tumorigenicity in rhabdomyosarcoma cells. J Biol Chem 2015;290:310-24. [Crossref] [PubMed]
- Frampton GM, Ali SM, Rosenzweig M, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov 2015;5:850-9. [Crossref] [PubMed]
- Dewaele M, Tabaglio T, Willekens K, et al. Antisense oligonucleotide-mediated MDM4 exon 6 skipping impairs tumor growth. J Clin Invest 2016;126:68-84. [Crossref] [PubMed]
- Comiskey DF Jr, Jacob AG, Singh RK, et al. Splicing factor SRSF1 negatively regulates alternative splicing of MDM2 under damage. Nucleic Acids Res 2015;43:4202-18. [Crossref] [PubMed]
- Shen S, Wang Y, Wang C, et al. SURVIV for survival analysis of mRNA isoform variation. Nat Commun 2016;7:11548. [Crossref] [PubMed]
- Griffith M, Griffith OL, Mwenifumbo J, et al. Alternative expression analysis by RNA sequencing. Nat Methods 2010;7:843-7. [Crossref] [PubMed]
- Zong Z, Li H, Yi C, et al. Genome-Wide Profiling of Prognostic Alternative Splicing Signature in Colorectal Cancer. Front Oncol 2018;8:537. [Crossref] [PubMed]
- Xiong Y, Deng Y, Wang K, et al. Profiles of alternative splicing in colorectal cancer and their clinical significance: A study based on large-scale sequencing data. EBioMedicine 2018;36:183-95. [Crossref] [PubMed]
- Mao S, Li Y, Lu Z, et al. Survival-associated alternative splicing signatures in esophageal carcinoma. Carcinogenesis 2019;40:121-30. [Crossref] [PubMed]
- Lin P, He RQ, Ma FC, et al. Systematic Analysis of Survival-Associated Alternative Splicing Signatures in Gastrointestinal Pan-Adenocarcinomas. EBioMedicine 2018;34:46-60. [Crossref] [PubMed]
- Li Y, Sun N, Lu Z, et al. Prognostic alternative mRNA splicing signature in non-small cell lung cancer. Cancer Lett 2017;393:40-51. [Crossref] [PubMed]
- Huang ZG, He RQ, Mo ZN. Prognostic value and potential function of splicing events in prostate adenocarcinoma. Int J Oncol 2018;53:2473-87. [PubMed]
- Ryan M, Wong WC, Brown R, et al. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res 2016;44:D1018-22. [Crossref] [PubMed]
- Ryan MC, Cleland J, Kim R, et al. SpliceSeq: a resource for analysis and visualization of RNA-Seq data on alternative splicing and its functional impacts. Bioinformatics 2012;28:2385-7. [Crossref] [PubMed]
- Lex A, Gehlenborg N, Strobelt H, et al. UpSet: Visualization of Intersecting Sets. IEEE Trans Vis Comput Graph 2014;20:1983-92. [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Li T, Wernersson R, Hansen RB, et al. A scored human protein-protein interaction network to catalyze genomic interpretation. Nat Methods 2017;14:61-4. [Crossref] [PubMed]
- Piva F, Giulietti M, Burini AB, et al. SpliceAid 2: a database of human splicing factors expression data and RNA target motifs. Hum Mutat 2012;33:81-5. [Crossref] [PubMed]
- Wagner GP, Kin K, Lynch VJ. A model based criterion for gene expression calls using RNA-seq data. Theory Biosci 2013;132:159-64. [Crossref] [PubMed]
- Chung IF, Chen CY, Su SC, et al. DriverDBv2: a database for human cancer driver gene research. Nucleic Acids Res 2016;44:D975-9. [Crossref] [PubMed]
- Li Q, Zheng S, Han A, et al. The splicing regulator PTBP2 controls a program of embryonic splicing required for neuronal maturation. Elife 2014;3:e01201. [Crossref] [PubMed]
- Sebastian S, Faralli H, Yao Z, et al. Tissue-specific splicing of a ubiquitously expressed transcription factor is essential for muscle differentiation. Genes Dev 2013;27:1247-59. [Crossref] [PubMed]
- Jacob AG, O'Brien D, Singh RK, et al. Stress-Induced Isoforms of MDM2 and MDM4 Correlate with High-Grade Disease and an Altered Splicing Network in Pediatric Rhabdomyosarcoma. Neoplasia 2013;15:1049-63. [Crossref] [PubMed]
- Comiskey DF Jr, Jacob AG, Sanford BL, et al. A novel mouse model of rhabdomyosarcoma underscores the dichotomy of MDM2-ALT1 function in vivo. Oncogene 2018;37:95-106. [Crossref] [PubMed]
- Sanchez G, Bittencourt D, Laud K, et al. Alteration of cyclin D1 transcript elongation by a mutated transcription factor up-regulates the oncogenic D1b splice isoform in cancer. Proc Natl Acad Sci U S A 2008;105:6004-9. [Crossref] [PubMed]
- Nowak DG, Amin EM, Rennel ES, et al. Regulation of vascular endothelial growth factor (VEGF) splicing from pro-angiogenic to anti-angiogenic isoforms: a novel therapeutic strategy for angiogenesis. J Biol Chem 2010;285:5532-40. [Crossref] [PubMed]
- Scheres SH, Nagai K. CryoEM structures of spliceosomal complexes reveal the molecular mechanism of pre-mRNA splicing. Curr Opin Struct Biol 2017;46:130-9. [Crossref] [PubMed]
- Galej WP. Structural studies of the spliceosome: past, present and future perspectives. Biochem Soc Trans 2018;46:1407-22. [Crossref] [PubMed]
- Smith DJ, Konarska MM, Query CC. Insights into branch nucleophile positioning and activation from an orthogonal pre-mRNA splicing system in yeast. Mol Cell 2009;34:333-43. [Crossref] [PubMed]
- Schellenberg MJ, Dul EL, MacMillan AM. Structural model of the p14/SF3b155. branch duplex complex. RNA 2011;17:155-65. [Crossref] [PubMed]
- Eke I, Cordes N. Focal adhesion signaling and therapy resistance in cancer. Semin Cancer Biol 2015;31:65-75. [Crossref] [PubMed]
- Tomar S, Plotnik JP, Haley J, et al. ETS1 induction by the microenvironment promotes ovarian cancer metastasis through focal adhesion kinase. Cancer Lett 2018;414:190-204. [Crossref] [PubMed]
- Luo M, Guan JL. Focal adhesion kinase: a prominent determinant in breast cancer initiation, progression and metastasis. Cancer Lett 2010;289:127-39. [Crossref] [PubMed]
- Zhao J, Guan JL. Signal transduction by focal adhesion kinase in cancer. Cancer Metastasis Rev 2009;28:35-49. [Crossref] [PubMed]
- Zhou B, Wang GZ, Wen ZS, et al. Somatic Mutations and Splicing Variants of Focal Adhesion Kinase in Non-Small Cell Lung Cancer. J Natl Cancer Inst 2018. [Crossref] [PubMed]
- Yao L, Li K, Peng W, et al. An aberrant spliced transcript of focal adhesion kinase is exclusively expressed in human breast cancer. J Transl Med 2014;12:136. [Crossref] [PubMed]
- Despeaux M, Chicanne G, Rouer E, et al. Focal adhesion kinase splice variants maintain primitive acute myeloid leukemia cells through altered Wnt signaling. Stem Cells 2012;30:1597-610. [Crossref] [PubMed]
- Rot S, Taubert H, Bache M, et al. Low HIF-1alpha and low EGFR mRNA Expression Significantly Associate with Poor Survival in Soft Tissue Sarcoma Patients; the Proteins React Differently. Int J Mol Sci 2018. [Crossref] [PubMed]
- Smolle MA, Leithner A, Posch F, et al. MicroRNAs in Different Histologies of Soft Tissue Sarcoma: A Comprehensive Review. Int J Mol Sci 2017. [Crossref] [PubMed]
- Yang Z, Li X, Yang Y, et al. Long noncoding RNAs in the progression, metastasis, and prognosis of osteosarcoma. Cell Death Dis 2016;7:e2389. [Crossref] [PubMed]
- Min L, Garbutt C, Tu C, et al. Potentials of Long Noncoding RNAs (LncRNAs) in Sarcoma: From Biomarkers to Therapeutic Targets. Int J Mol Sci 2017. [Crossref] [PubMed]
- de Almeida SF, Carmo-Fonseca M. Design principles of interconnections between chromatin and pre-mRNA splicing. Trends Biochem Sci 2012;37:248-53. [Crossref] [PubMed]
- Wada M, Horinaka M, Yasuda S, et al. PDK1 is a potential therapeutic target against angiosarcoma cells. J Dermatol Sci 2015;78:44-50. [Crossref] [PubMed]
- Le Gallo M, Rudd ML, Urick ME, et al. The FOXA2 transcription factor is frequently somatically mutated in uterine carcinosarcomas and carcinomas. Cancer 2018;124:65-73. [Crossref] [PubMed]
- Joseph CG, Hwang H, Jiao Y, et al. Exomic analysis of myxoid liposarcomas, synovial sarcomas, and osteosarcomas. Genes Chromosomes Cancer 2014;53:15-24. [Crossref] [PubMed]
- Chudasama P, Renner M, Straub M, et al. Targeting Fibroblast Growth Factor Receptor 1 for Treatment of Soft-Tissue Sarcoma. Clin Cancer Res 2017;23:962-73. [Crossref] [PubMed]
- Sveen A, Kilpinen S, Ruusulehto A, et al. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 2016;35:2413-27. [Crossref] [PubMed]
- Kovac M, Blattmann C, Ribi S, et al. Exome sequencing of osteosarcoma reveals mutation signatures reminiscent of BRCA deficiency. Nat Commun 2015;6:8940. [Crossref] [PubMed]


