
Impact of negative lymph nodes on colon cancer survival and exploring relevant transcriptomics differences through real-world data analyses
Introduction
The prognostic parameters of post-surgery colon cancer have been identified, but the multivariate predictive model of survival for individual patients is still unclear. The number of lymph nodes harvested and examined carries with it certain prognostic and therapeutic implications (1). Some studies have identified the number of negative lymph nodes (NLNs) as positively related to survival, but the findings have been inconsistent, and the underlying mechanisms have not been clarified (2-4).
NLN was reported as a prognostic factor in IIIB/IIIC colon cancer (2). The number of assessed lymph nodes was positively associated with survival in T3N0 colon cancer and Dukes’ B colorectal cancer (5,6), which thus verified the prognostic role of NLN number. The NLN may represent an independent prognostic factor apart from tumor stage. However, these studies were conducted on surgeries performed before 2000 and lack a paralleled update. A recent study showed that number of lymph nodes harvested had no prognostic impact on node-negative rectal cancers treated with neoadjuvant therapy (3).
Comparatively, little research has been devoted to elucidating the molecular mechanisms in different NLN subgroups in the genome. In the present study, we hypothesized that NLN would offer extra precision in predicting the outcome of colon cancer and establish a predictive nomogram to provide visualization information. To reveal the heterogeneity affecting NLN, we conducted analyses of two large databases, the Surveillance, Epidemiology, and End Results (SEER) and the Cancer Genome Atlas (TCGA) databases, which may represent real-world conditions more accurately.
Methods
Patients and database
SEER database
Data were obtained from the SEER Program (www.seer.cancer.gov) with the following search parameters: SEER*Stat Database: Incidence-SEER 18 Regs Research Data, Nov. 2017 Sub (1973–2015), released June 3rd, 2018. Patients with pathological diagnosis of colon cancer as the first primary cancer from 2000 to 2015 based on the International Classification of Disease for Oncology, Third Edition (histology code: ICD-O-3/WHO 2008), and site recode “colon excluding rectum”, were enrolled, while patients diagnosed before 2000 were excluded. Other exclusion criteria were as follows: patients with missing data on lymph node dissection, or unclear data for cancer-specific survival (CSS).
The size of the primary tumor was measured from the pathology report, operative report, endoscopic examination, and the radiographic report, in priority order. The surgery of the primary site included local tumor destruction, local tumor excision, partial colectomy segmental resection, subtotal colectomy/hemicolectomy, total colectomy, total proctocolectomy, colectomy or coloproctotectomy with resection of the contiguous organ(s) (NOS), and colectomy (NOS).
Clinical data on the regional lymph node examined, regional positive lymph node, sex, tumor grade, TNM stage, age at diagnosis, year at diagnosis, survival months, and survival status were extracted. Regional lymph nodes for all colon subsites, including epicolic (next to the bowel wall), paracolic/pericolic, colic (NOS), and nodule(s) in pericolic fat, were also collected. Regional nodes examined refers to the total number of regional lymph nodes that were removed and examined by the pathologist. Regional nodes positive records the exact number of regional lymph nodes examined by the pathologist that were found to contain metastases. NLN was defined as regional nodes examined minus regional nodes positive. For CSS, death from cancer is an event, and for overall survival (OS), any death is an event.
TCGA database
TCGA colon adenocarcinoma (COAD) dataset was obtained on June 9th, 2018. The clinical parameters and mRNA expression values were downloaded from the TCGA data portal (https://portal.gdc.cancer.gov). Patients with detailed information of lymphadenectomy and survival status were included. Patients with missing data on lymph node dissection were excluded.
Ethical approval was approved by the Shandong Provincial Qianfoshan Hospital review board. Informed consents from patients were waived due to the anonymity of individual patient data.
Assessments of molecular features
The six infiltrated immune cell subgroups including B cells, CD4+ T cells (T CD4), CD8+ T cells (T CD8), neutrophils (Neu), macrophages (Mac) and dendritic cells (DC) were available at a user-interactive website (http://cistrome.org/TIMER). Microsatellite instability (MSI) status and hypermutation were obtained from The Cancer Immunome Atlas (TCIA, https://tcia.at/).
The gene expression data were downloaded from the TCGA Data Portal. The RNA-seq was summarized by read count, and was normalized for differential analyses performed using edgeR package. Fold change >2 was adopted as the threshold to screen for differentially expressed genes (DEGs). These DEGs were further analyzed using Metascape (http://metascape.org). Protein-protein interaction (PPI) and network analyses of the DEGs, were performed using the following databases: BioGrid (7), InWeb_IM (8), and OmniPath (9). Molecular Complex Detection (MCODE) algorithm was adopted to screen densely connected modules (10), while Bubble Chart was plotted using the OmicShare tools, a free online platform for data analysis (www.omicshare.com/tools).
Statistical methods
The optimal cutoff of NLNs number was calculated by receiver operating characteristic (ROC) curve to predict the outcome. Baseline categorical variables and continuous variables were assessed by χ2 test and Mann-Whitney U-test, respectively. Kaplan-Meier methodology was used to assess the influence of NLNs on CSS and OS. Hazard ratios (HRs) and 95% confidence intervals (CIs) were measured by multivariate Cox proportional hazard models with a stepwise forward method.
We further constructed a nomogram based on the multivariate Cox regression results of the training cohort (SEER), and patients from the TCGA database were included as a test cohort. The Harrell’s C-index was used to quantify the discrimination performance of the nomogram. Calibration curves with 1,000-resample bootstrap validation at different time points were generated to depict the calibration of each model. External validation was performed by applying the nomogram to the test group using the TCGA dataset.
All statistical tests were two-sided, and a P value <0.05 was judged to be statistically significant. Statistical analyses were conducted using R (V.3.5.0, the R Foundation for Statistical Computing, Vienna, Austria) or SPSS 20.0 (SPSS Inc., Chicago, IL, USA) software. The forest plot was performed with Stata Statistical Software (Version 12.0).
Results
Cohort
A total of 314,398 patients from the SEER database were enrolled in this study. Of all patients, 262,003 (83.3%) underwent lymphadenectomy, and 52,395 (16.7%) were confirmed without lymphadenectomy. In total, 154,581 (49.2%) were males and 159,817 (50.8%) were females. The median age of patients included was 69 years (4–108 years). The baseline characteristics of the SEER cohort are shown in Table 1.
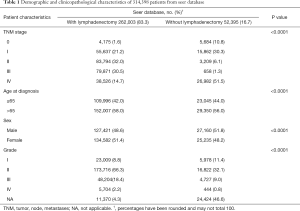
Full table
A total of 433 patients from TCGA COAD data with lymphadenectomy information were used as validation data and for further mechanism analysis. Clinicopathological features of the 433 patients in the validation cohort are listed in Table 2.

Full table
Impact of NLN number on survival
Firstly, we examined the influence of the lymphadenectomy on the whole cohort and found lymphadenectomy to be associated with better survival in both CSS and OS (Figure 1A,B, both P<0.0001).
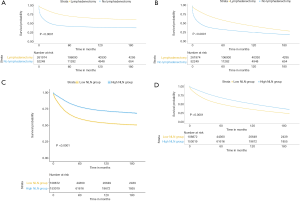
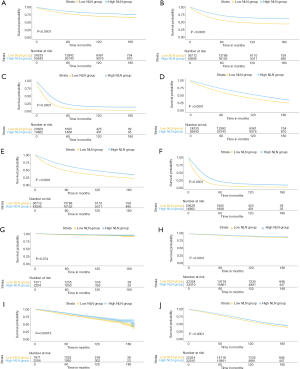
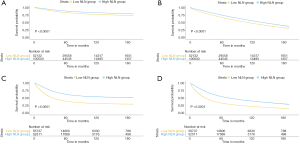
We further analyzed the prognostic effect of NLN in the cohort who underwent lymphadenectomy. The median value of the NLN was 13 (range, 0–90). We determined the best cut-off value by ROC and divided the whole cohort into two subgroups (low NLN, 0–11, and high NLN, ≥12). Kaplan-Meier analyses showed that NLNs was positively associated with CSS and OS (Figure 1C,D, both P<0.0001). The survival benefit of the NLNs appeared to be stronger in stage II–IV (Figure 2A,B,C for CSS and Figure 2D,E,F for OS, all P<0.0001) compared to stage 0–I (Figure 2G,H for CSS and Figure 2I,J for OS, P=0.074, P<0.0001, P=0.00013, and P<0.0001, respectively). These survival benefits were constant regardless of lymph node status, and survival advantages were shown in both negative and positive lymph node subgroups (Figure 3A,B,C,D, all P<0.0001).
The mean NLN and number of lymph nodes examined both increased as the date of diagnosis increased. However, very little change was shown in the number of positive lymph nodes (Figure 4A). A significant correlation was identified between the number of lymph nodes retrieved and NLN (r=0.931, P<0.0001), while the correlations of the positive lymph nodes and NLN (r=−0.197, P<0.0001), and the lymph nodes retrieved and positive lymph nodes (r=0.175, P<0.0001) were weak or negligible.
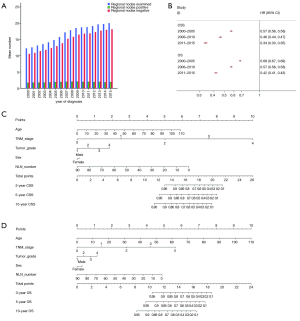
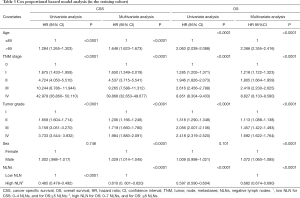
The results of univariate and multivariate Cox proportional-hazards model analyses are shown in Table 3. A univariate analysis per subset of 5 years showed that NLN was positively associated with survival consistently despite diagnosis time (Figure 4B). Multivariate analyses confirmed that patients with a high NLN had improved CSS compared to a low NLN (HR =0.610, 95% CI, 0.601–0.620, P<0.0001). This trend remained consistent for OS (HR =0.682, 95% CI, 0.674–0.690, and P<0.0001 (high vs. low)). A higher NLN was related to better survival in colon cancer, independent of sex, age, tumor grade, and TNM stage (Table 3).
Full table
Predictive nomograms for mortality
We then constructed nomograms based on the final multivariate analyses for the training cohort (Figure 4C for CSS and Figure 4D for OS) at 3 years, 5 years, and 10 years. In the nomogram panels, the first row is the point assigned for each variable. The sum of the points for each variable equals the total points. A vertical line drawn from this point can obtain the 3-, 5-, and 10-year probability of survival. The nomogram was externally validated by using 433 patients from TCGA COAD data. Tumor grade and CSS were not available in TCGA cohort, so our findings were validated in the test cohort for OS without differentiation.
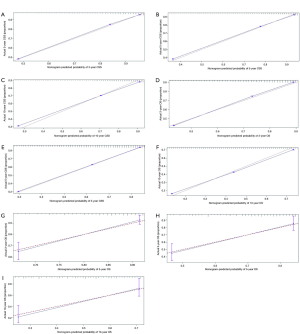
The c-statistics of these predictive models were 0.790 (95% CI, 0.788–0.792) for CSS, 0.734 (95% CI, 0.732–0.736) for OS in the training cohort, and 0.743 (95% CI, 0.688–0.798) for OS in the test cohort, which indicates a good ability to predict outcome. The calibration plots showed good agreement of the prediction and observation in survival (Figure S1A,B,C for CSS, Figure S1D,E,F for OS in the training cohorts and Figure S1G,H,I for OS in the test cohort).
Molecular features analyses
To explore the difference of global gene expression levels, we divided TCGA COAD database into high (≥12 NLN) and low (0–11 NLN) groups based on the cut-off obtained from the SEER database.
Most immune cells infiltrated did not differ between the high and low NLN groups (Figure 5A, most P>0.05) except B cell (P=0.002) and Mac (P<0.0001). Less B cell and Mac infiltration was observed in the high NLN group. High NLN tumor displayed markedly higher frequencies of high MSI (68/329, 20.669%) than the low NLN group (5/88, 5.682%) with OR =4.325 (95% CI, 1.687–11.085 and P=0.001). Meanwhile, the frequency of hypermutation was higher in the high NLN group (54/238, 22.689%) compared to the low NLN group (5/78, 6.410%) with OR =4.285 (95% CI, 1.648–11.140 and P=0.001). Therefore, diverse biological features may be present in different NLN groups.
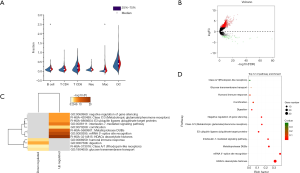
RNA-seq files of these colon cancer patients (362 in the high NLN group and 92 in the low NLN group) were downloaded from TCGA. EdgeR analysis recognized 1,818 DEGs in high NLN compared with low NLN. Most of these genes showed more upregulation [1760] than downregulation (58) (Figure 5B).
Enrichment analysis
The Metascape online tool was used for enrichment analysis (Figure 5C). Enriched terms were represented as a dynamic bubble chart (Figure 5D). These analyses revealed the top 12 modules with their typical enriched terms which were related to histone modifiers, mRNA splicing, metalloprotease DUB, etc. (Table S1). These relationships were further described using Metascape network analysis (Figure 6A), which showed the most significant DEGs were involved in the regulation of histones and histone deacetylases (HDACs).

Full table
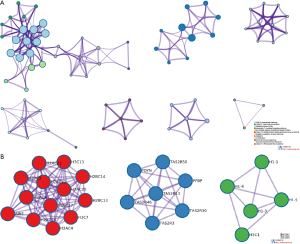
PPIs are of great importance to most biological processes. The PPI network and MCODE components by all DEGs are shown in Figure 6B. MCODE networks identified the top most densely connected network components by cluster related to 3 MCODE. MCODE1 is involved in HDAC deacetylate histones (log10P=−28.1), systemic lupus erythematosus (log10P=−26.2), and histone acetyltransferases (HATs) acetylate histones (log10P=−25.9). MCODE 2 is involved in Class C/3 (metabotropic glutamate/pheromone receptors) (log10P=−12.8), detection of chemical stimulus involved in sensory perception of bitter taste (log10P=−12.6), and G alpha (i) signaling events(log10P=−12.4). MCODE 3 is involved in apoptosis-induced DNA fragmentation(log10P=−12.6), the formation of senescence-associated heterochromatin foci (SAHF) (log10P=−12.2), and nucleosome positioning (log10P=−12.1). Specifically, the HDACs deacetylate histones MCODE was found to be obviously up-regulated in the high NLN compared to the low NLN group with the best-scoring terms by P value. These results suggest agents targeting histones may be a good therapeutic choice for the high NLN group.
Discussion
Our study demonstrated that NLN was a significant predictive variable for survival in a large cohort of colon cancer patients, and, for the first time, established a nomogram that includes NLN based on real-world analyses. This nomogram was externally validated by TCGA database and showed good agreement with the prediction and observation in survival. We identified high NLN as being associated with less B cell and macrophage infiltration, high MSI, and hypermutation. We also found that histone modifiers were the most significant different biological process between the two groups.
The correlation of lymph nodes and survival could be confounded by the prognostic value of increasing positive lymph node number, and the establishment of the prognostic effect of NLN explained these inconsistent findings. Our study showed that as the number of lymph nodes examined increased, so did the NLN number; meanwhile, the positive lymph node number did not markedly increase, which could mean that it is possible that dissection of NLNs itself improved the patients’ outcome.
There may be divergent opinions concerning the prognostic role of NLN. Firstly, different views exist relating to the diagnostic or therapeutic role of lymphadenectomy. On the one hand, higher NLN reflects more extensive lymph node detection. A significant positive correlation was observed between the NLNs and lymph nodes examined (11), which was also demonstrated by our study. The most current view on the dissection of lymph nodes is that it is diagnostic. One paradigm assumes that cancer is a systemic disease involving complex interactions of host and tumor at inception, and lymphadenectomy itself does not improve survival (12). The assessment of high lymph nodes examined allows for risk reduction of understaging in colorectal cancer which decreases misclassification of node-positive patients as node-negative (2,13-15). The correct staging is the basis of the optimum strategy for adjuvant therapy, while understaged patients might miss the opportunity of getting adjuvant therapies, resulting in poor outcome (16-18). On the other hand, few studies have found that lymphadenectomy can have a therapeutic effect (19). Under Halsted’s concept, lymphadenectomy should provide both staging and therapeutic roles (12). Indirect evidence was inferred from the analysis of intergroup trial INT-0089. The trial indicated that survival benefit was achieved as the number of lymph nodes retrieved increased when the number of lymph nodes involved was controlled and even when no nodes were involved (20). Secondly, the number of lymph node retrieved may also be influenced by the adequacy of the surgeon (20) and pathologist. A high quality of the examination of the specimen by the pathologist may result in more positive lymph nodes being found and more accurate staging (21). Thirdly, lymph nodes are suggested to be an underlying factor affecting antitumor immune response. The tumor might induce an inflammatory response which influences the number of lymph nodes that are found, as they can be palpated more easily by the pathologist. Furthermore, our research identified DEGs in different groups, which have potential as genetic factors that can also predict improved survival.
As previously reported, MSI-high and high inflammatory cell infiltration were associated with the retrieval of a larger number of lymph nodes (22). MSI-H tumor was characterized by containing more tumor-infiltrating lymphocytes (23). However, most of the infiltrated inflammatory cells did not differ in high and low NLN groups in this study despite the relationship between high NLN and MSI-H. The difference here may be explained as a result of the potential differences of lymph nodes retrieved and NLNs, the different detection methods, and the diverse populations. The relationship between MSI and the number of NLNs may be simply caused by the immunological response and thus the fact that lymph nodes may be palpated more easily because of their inflammatory reaction. Our study also found that high B cell and Mac infiltration were observed in the low NLN group, which could be partly supported by recent research that Macs could uptake cancer cell exosomes and promote the formation of the lymphatic network in sentinel lymph nodes (24). Despite these findings, higher levels of evidence are lacking.
Lymph node status is a vital criterion of demarcation for M0 disease, while M1 disease is categorized as stage IV regardless of N status. Palliative resection has often been performed in advanced disease to alleviate symptoms, to enhance life quality, and to prevent complications with limited lymph node retrieved. Our study demonstrated that NLN was also associated with a survival benefit in the M1 stage, which could be observed in the subgroup analysis of stage IV disease. This was consistent with the findings in stage IV gastric cancer, and in metastatic colorectal cancer (25). Therefore, the prognostic role of N status has been underestimated in metastasis disease. Meanwhile, we should carefully select the advanced patients to maximize the effect of lymphadenectomy and minimize morbidity by a multidisciplinary strategy.
Our study shows that an increased NLN number reflects histone modifiers differential expression for colon cancer for the first time. The HDACs and HATs that can regulate transcription by modifying the deacetylating and acetylating state of histones, which may further trigger many nuclear events. Recent evidence suggests that a shift in the balance of acetylase and deacetylase activity plays an important role in the pathogenesis of cancer (26). Decreased levels of histone acetylation have been associated with poorer survival (27). This was consistent with our finding that the low NLN group expressed down-regulated histone genes which was correlated with poor outcome. Meanwhile, the high NLN group may be more sensitive to HDAC inhibitors. A more comprehensive understanding of the underlying mechanism may lead to novel therapeutic interventions for colon cancer.
Some limitations should be considered when interpreting our results. Considering its retrospective nature, more detailed clinical information such as the details of chemotherapy are unavailable, which might have affected the number of NLNs and be a contributor of bias in this study. Also, the data’s lack of molecular subtypes might have concealed significant prognostic information from researchers.
Conclusions
In conclusion, our study established the prognostic role of NLN in colon cancer and constructed a simple nomogram to estimate CSS and OS. We postulated that the high NLN group may represent a biological subtype with histone modifier gene enriched expression, and patients with high NLN number may be more sensitive to HDAC inhibitor. Further studies about high NLN are needed to determine the template of lymph node dissection, the underlying molecular mechanism, and the feasibility of targeting histone in colon cancer.
Acknowledgments
We gratefully acknowledge The Cancer Genome Atlas (TCGA) Research Network and The Cancer Immunome Database (TCIA) for providing the primary data, the TIMER web server for providing a comprehensive resource for systematic analysis of immune infiltrates, and the SEER program tumor registries for the creation of the SEER-Medicare database.
Funding: This work was supported by the National Natural Science Foundation of China (grant no. 81672974, and no. 81602719) and Science and Technology Development Plan of Shandong Province (No. 2017GSF18111).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Shandong Provincial Qianfoshan Hospital review board (No. S017). Informed consents from patients were waived due to the anonymity of individual patient data.
References
- Nelson H, Petrelli N, Carlin A, et al. Guidelines 2000 for colon and rectal cancer surgery. J Natl Cancer Inst 2001;93:583-96. [Crossref] [PubMed]
- Johnson PM, Porter GA, Ricciardi R, et al. Increasing negative lymph node count is independently associated with improved long-term survival in stage IIIB and IIIC colon cancer. J Clin Oncol 2006;24:3570-5. [Crossref] [PubMed]
- Degiuli M, Arolfo S, Evangelista A, et al. Number of lymph nodes assessed has no prognostic impact in node-negative rectal cancers after neoadjuvant therapy. Results of the "Italian Society of Surgical Oncology (S.I.C.O.) Colorectal Cancer Network" (SICO-CCN) multicentre collaborative study. Eur J Surg Oncol 2018;44:1233-40. [Crossref] [PubMed]
- Zhu Z, Chen H, Yu W, et al. Number of negative lymph nodes is associated with survival in thoracic esophageal squamous cell carcinoma patients undergoing three-field lymphadenectomy. Ann Surg Oncol 2014;21:2857-63. [Crossref] [PubMed]
- Swanson RS, Compton CC, Stewart AK, et al. The prognosis of T3N0 colon cancer is dependent on the number of lymph nodes examined. Ann Surg Oncol 2003;10:65-71. [Crossref] [PubMed]
- Caplin S, Cerottini JP, Bosman FT, et al. For patients with Dukes' B (TNM Stage II) colorectal carcinoma, examination of six or fewer lymph nodes is related to poor prognosis. Cancer 1998;83:666-72. [Crossref] [PubMed]
- Stark C, Breitkreutz BJ, Reguly T, et al. BioGRID: a general repository for interaction datasets. Nucleic Acids Res 2006;34:D535-9. [Crossref] [PubMed]
- Li T, Wernersson R, Hansen RB, et al. A scored human protein-protein interaction network to catalyze genomic interpretation. Nat Methods 2017;14:61-4. [Crossref] [PubMed]
- Turei D, Korcsmáros T, Saez-Rodriguez J. OmniPath: guidelines and gateway for literature-curated signaling pathway resources. Nat Methods 2016;13:966-7. [Crossref] [PubMed]
- Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003;4:2. [Crossref] [PubMed]
- Li Q, Zhuo C, Cai G, et al. Increased number of negative lymph nodes is associated with improved cancer specific survival in pathological IIIB and IIIC rectal cancer treated with preoperative radiotherapy. Oncotarget 2014;5:12459-71. [PubMed]
- Fisher B. From Halsted to prevention and beyond: advances in the management of breast cancer during the twentieth century. Eur J Cancer 1999;35:1963-73. [Crossref] [PubMed]
- Pheby DF, Levine DF, Pitcher RW, et al. Lymph node harvests directly influence the staging of colorectal cancer: evidence from a regional audit. J Clin Pathol 2004;57:43-7. [Crossref] [PubMed]
- Miller EA, Woosley J, Martin CF, et al. Hospital-to-hospital variation in lymph node detection after colorectal resection. Cancer 2004;101:1065-71. [Crossref] [PubMed]
- Joseph NE, Sigurdson ER, Hanlon AL, et al. Accuracy of determining nodal negativity in colorectal cancer on the basis of the number of nodes retrieved on resection. Ann Surg Oncol 2003;10:213-8. [Crossref] [PubMed]
- Wong JH, Severino R, Honnebier MB, et al. Number of nodes examined and staging accuracy in colorectal carcinoma. J Clin Oncol 1999;17:2896-900. [Crossref] [PubMed]
- Schrag D, Gelfand SE, Bach PB, et al. Who gets adjuvant treatment for stage II and III rectal cancer? Insight from surveillance, epidemiology, and end results--Medicare. J Clin Oncol 2001;19:3712-8. [Crossref] [PubMed]
- Oliveria SA, Yood MU, Campbell UB, et al. Treatment and referral patterns for colorectal cancer. Med Care 2004;42:901-6. [Crossref] [PubMed]
- Ong ML, Schofield JB. Assessment of lymph node involvement in colorectal cancer. World J Gastrointest Surg 2016;8:179-92. [Crossref] [PubMed]
- Le Voyer TE, Sigurdson ER, Hanlon AL, et al. Colon cancer survival is associated with increasing number of lymph nodes analyzed: a secondary survey of intergroup trial INT-0089. J Clin Oncol 2003;21:2912-9. [Crossref] [PubMed]
- Sigurdson ER. Lymph node dissection: is it diagnostic or therapeutic? J Clin Oncol 2003;21:965-7. [Crossref] [PubMed]
- Kim YW, Jan KM, Jung DH, et al. Histological inflammatory cell infiltration is associated with the number of lymph nodes retrieved in colorectal cancer. Anticancer Res 2013;33:5143-50. [PubMed]
- Phillips SM, Banerjea A, Feakins R, et al. Tumour-infiltrating lymphocytes in colorectal cancer with microsatellite instability are activated and cytotoxic. Br J Surg 2004;91:469-75. [Crossref] [PubMed]
- Sun B, Zhou Y, Fang Y, et al. Colorectal cancer exosomes induce lymphatic network remodeling in lymph nodes. Int J Cancer 2019;145:1648-59. [Crossref] [PubMed]
- Zhuo C, Ying M, Lin R, et al. Negative lymph node count is a significant prognostic factor in patient with stage IV gastric cancer after palliative gastrectomy. Oncotarget 2017;8:71197-205. [Crossref] [PubMed]
- Ishihara S, Hayama T, Yamada H, et al. Prognostic impact of primary tumor resection and lymph node dissection in stage IV colorectal cancer with unresectable metastasis: a propensity score analysis in a multicenter retrospective study. Ann Surg Oncol 2014;21:2949-55. [Crossref] [PubMed]
- Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer 2006;6:38-51. [Crossref] [PubMed]


