Association between TIM-3 polymorphisms and cancer risk: a meta-analysis
Introduction
T-cell immunoglobulin- and mucin-domain-containing molecule 3 (TIM-3), also known as hepatitis A virus cellular receptor 2 (HAVCR2), is a T-helper 1 (Th1)-specific cell surface protein and functions to regulate an immune checkpoint by activating macrophages and enhancing experimental autoimmune encephalomyelitis severity (1,2). TIM-3 also served as a specific cell surface marker for Th1 CD4+ T lymphocytes, CD8+ T lymphocytes, dendritic cells, Th17 lymphocytes, but not for Th2 cells and is a member of the Ig super family (3-5). The Tim-3 pathway may interact with the programmed cell death 1 pathway in dysfunctional CD8+ T cells and Tregs in cancers (2). Molecularly, TIM-3 interactions with its ligand, galectin-9, will negatively regulate tumor infiltrating CD4+ T and CD8+ T cells via the selective elimination of TIM-3-positive T cells to control T lymphocyte populations and activities (6,7). Functionally, TIM-3-positive CD4+ and CD8+ T cells reduce the production and secretion of cytokines and/or show less proliferation after exposure to tumor antigens (7). In contrast, the inhibition of TIM-3 expression or activity was shown to restore antigen-induced specific cell proliferation and cytokine levels (7). Thus, altered TIM-3 expression would be considered to be involved in cancer susceptibility. Notably, a recent meta-analysis demonstrated that higher TIM-3 expression was associated with advanced tumor stage and shorter overall survival of patients with various cancers, including bladder cancer, non-small cell lung cancer, gastric cancer, ovarian cancer, cervical cancer, hepatitis B virus-associated hepatocellular carcinoma, and clear cell renal cell carcinoma (8). Targeting of TIM-3 was considered in a novel cancer immunotherapy strategy (9). In this regard, the assessment of TIM-3 alteration and activity could help us understand the role of TIM-3 in cancer susceptibility.
Human TIM-3 is localized at chromosome 5q33.3, which contains a large number of single nucleotide polymorphisms (SNPs) (10). These TIM-3 polymorphisms were reported to associate with TIM-3 expression and activity and then modify cancer risk in various populations (11). Notably, a previous meta-analysis (12) assessed TIM-3 polymorphisms (−1516G/T, −574G/T, and +4259T/G) and showed an association with increased cancer risk. TIM-3 promoter region polymorphisms (−1516G/T, −882C/T, and −574G/T) significantly induced genetic susceptibility of gastric cancer (13), and TIM-3 −574G/T polymorphism was associated with a risk of developing myasthenia gravis-associated thymoma (14). Moreover, TIM-3 rs10053538 also increased breast cancer susceptibility and promoted breast cancer progression (11). We performed this meta-analysis study to better understand and precisely identify TIM-3 SNPs and to associate them with cancer risk. We expect to provide more insightful information and to support TIM-3 SNPs as biomarkers in predicting cancer susceptibility.
Methods
Literature search to identify eligible and relevant studies
We searched literature for all published studies that assessed an association between TIM-3 polymorphism and cancer in PubMed, EMBASE, China Biology Medical Literature Database (CBM), Wanfang Data, and the China National Knowledge Infrastructure (CNKI) (up to July 10, 2018). TIM-3 is also known as “CD366, HAVCR2, KIM-3, TIM3, TIMD-3, TIMD3, or T-cell immunoglobulin mucin-3”; thus, our search also included these words as keywords in addition to “polymorphism or single nucleotide” and “carcinoma or neoplasms”. Moreover, in the CBM, Wanfang, and CNKI databases, our search terms used these corresponding keywords in Chinese characters. Our literature search was only restricted to human studies. Next, we retrieved all eligible studies and checked their bibliographies for further relevant publications. If insufficient data were available in trial publications or for unpublished trials, we contacted the investigators to obtain the data. The inclusion criteria included (I) case-control or nested case-control studies focusing on the association between TIM-3 and cancer risk; (II) having adequate data to calculate the genotypic odd ratio (OR) and corresponding 95% confidence interval (CI), including total number of cancer cases and controls, as well as the number of cases and controls for each genotype; (III) all full text articles. The exclusion criteria were (I) publication of reviews, tutorials and letters; (II) animal studies; and (III) duplicate publications. However, when a similar or identical patient population was used in several publications, we only selected the most recent, largest, or complete study for our data analysis. If more than one ethnic population was enrolled in a study, each population was regarded as an independent study for our data analysis.
Data extraction and quality assessment
Data were extracted independently by two investigators (H Fang and M Sun) from all eligible studies, which included the first author’s name, year of publication, country, ethnicity of the population, tumor types, sample source, genotyping methods, matching criteria, genotype distribution and control source, Hardy-Weinberg equilibrium (HWE), and number of participants with each genotype in the cases and controls.
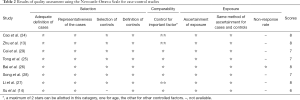
Quality assessment was also performed by two investigators (H Fang and M Sun) independently using the Newcastle-Ottawa Scale (NOS) according to a previous publication (15). The NOS contains eight items, which are categorized into three perspectives, i.e., selection of the study group and the comparability and exposure of the interest. In this study, we scored the quality of each study for a maximum of 1 point for each item in the selection and exposure perspectives and a maximum of 2 points in comparability. Any discrepancy was solved through discussion between the two investigators. If they could not reach a consensus, other investigators were consulted to resolve the discrepancy to make a final decision through a vote majority.
Statistical analysis
The HWE was estimated first in controls for each study using the χ2 test and a P value <0.05 was considered to indicate significant disequilibrium according to a previous study (16). The crude OR and 95% CI calculated using Woolf’s method were used to estimate the association between TIM-3 polymorphism and cancer susceptibility under the dominant, recessive, homozygous, heterozygous, and allelic models. The heterogeneity across all eligible comparisons was assessed using χ2-based Cochrane’s Q statistics (the significance level was set at P<0.10) according to a previous study (17) and the I2 statistics. The following thresholds were used to quantify the I2 metric: I2=0–25%, no heterogeneity; I2=25–50%, moderate heterogeneity; I2=50–75%, large heterogeneity; I2=75–100%, extreme heterogeneity (18). Furthermore, the data were combined using both fixed effect (Mantel-Haenszel) (19) and random effect (DerSimonian and Laird) (20) models. Unless stated otherwise, the random effect estimates were reported. Meta-regression analysis was applied to detect the source of heterogeneity. To explore the sources of in-between-study heterogeneity, we conducted a stratified analysis according to the source of control groups and the type of cancer.
To assess the impact of a single study on the pooled OR and to confirm the stability of the results, we performed a sensitivity analysis to repeat analyses by the sequential removal of individual studies (21). The Funnel plots and Egger’s test (22,23) were used to explore the presence of publication bias.
All P values were two-tailed, and P<0.05 was considered statistically significant. All statistical analyses were performed with STATA version 13.0 (Stata Corporation, College Station, TX, USA).
Regarding the haplotype association analysis, we utilized a multivariate analysis strategy with summary-based data and methods that used count data in a generalized linear mixed model framework (logistic regression).
Results
Study selection
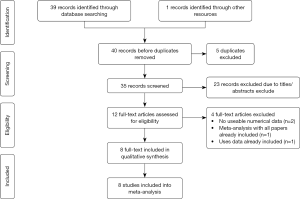
In this study, we identified a total of 40 studies (39 from PubMed and one from CBM and CNKI). After removing duplicated studies, we excluded 29 publications and abstracts and obtained 11 publications. While inspecting their full-text, we found that two studies were not relevant to TIM-3 SNPs association and that one study contained overlapping data; thus, these three studies were also excluded (21). Eight studies (13,14,24-29) remained, the data of which were incorporated into our systematic review and meta-analysis (Figure 1).
Characteristics of the included studies
All eligible publications were case-control studies with a total of 4,852 participants (2,229 cases and 2,623 controls). Seven publications were in English, and one was in Chinese (13). However, all studies were performed in China, and the genotype distributions in the controls met the HWE.
Moreover, all cancer patients were enrolled according to clinical examination and pathological evidence, whereas the control groups had no signs or symptoms of cancer. Polymerase chain reaction (PCR) was utilized to genotype the TIM-3 polymorphisms, and four TIM-3 SNPs and haplotypes were included in seven studies of −1516G/T (13,24-29), two studies of −882C/T (13,24), seven of −574G/T (13,14,24-26,28,29), five of +4259T/G (14,24-26,28,29), and four of haplotypes (25,26,28,29). The main characteristics of the included studies are shown in Table 1. Furthermore, all included studies were of high quality with a NOS score ≥6 (Table 2).

Full table
Full table
Meta-analysis
TIM-3 −1516G/T polymorphism
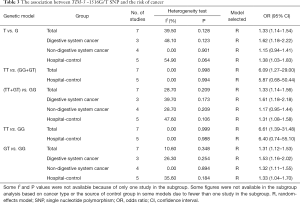
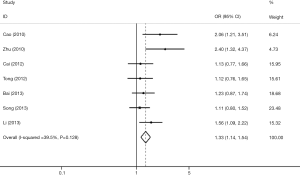
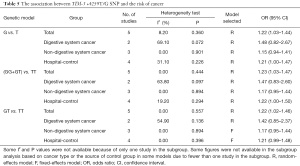
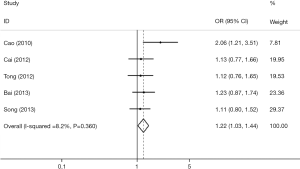
Seven studies investigated the association between TIM-3 −1516G/T and cancer risk in 2,229 cases and 2,623 control subjects (Table 3). There was significant between-study heterogeneity (P=0.128), and the value of the I2 index (I2=39.50%) and the pooled risk estimates indicated that the TIM-3 −1516G/T polymorphism was associated with an increased risk in developing cancers overall [T vs. G (Figure 2); GT vs. GG; TT vs. GG; (TT+GT) vs. GG; TT vs. (GT+GG)]. In the subgroup analyses based on cancer type, we found that the TIM-3 −1516G/T polymorphism was only associated with an increased cancer risk in the digestive system [T vs. G; GT vs. GG, TT vs. GG, (TT+GT) vs. GG; TT vs. (GT+GG)]. Furthermore, in the sensitivity analysis based on the source of controls (population control vs. hospital control), we found that the TIM-3 −1516G/T polymorphism was associated with an increased cancer risk in the populations of hospital origin [T vs. G; GT vs. GG; (TT+GT) vs. GG] (Table 3).
Full table
TIM-3 −882C/T polymorphism
Two studies assessed the association between TIM-3 −882C/T and cancer risk (13,24). We did not make a pooled estimate with only two studies. The data in the original two studies showed that subjects carrying the CT genotype and T allele had a significantly increased risk of cancer compared with those carrying the CC genotype and C allele, respectively (T vs. C, CT vs. CC, Figure S1, Table S1).
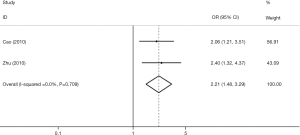
Full table
TIM-3 −574G/T polymorphism
Seven studies associated TIM-3 −574G/T with cancer susceptibility in 2,074 cases and 2,385 control subjects (Table 4). We found that subjects carrying the GT genotype or T allele had a significantly increased risk of overall cancer compared with those carrying the GG genotype or G allele, respectively (T vs. G; GT vs. GG). In the subgroup analyses based on cancer types, we found that the TIM-3 −574G/T polymorphism was not associated with any increased cancer risk from the digestive system (T vs. G; GT vs. GG) but was associated with an increased risk of non-digestive system cancer [T vs. G (Figure 3); GT vs. GG]. Moreover, in the sensitivity analyses based on the source of control, we found that the TIM-3 −574G/T polymorphism was associated with an increased cancer risk in the hospital population (T vs. G; GT vs. GG, Table 4).

Full table
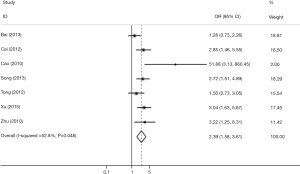
TIM-3 +4259T/G polymorphism
We retrieved five studies that determined the association between TIM-3 +4259 T/G and cancer risk in 1,868 cases and 2,566 control subjects (Table 5). The data showed that subjects carrying the GT genotype, GG+GT genotype, or G allele had a significantly increased risk of overall cancer compared with those carrying the TT genotype, TT genotype or T allele, respectively [G vs. T (Figure 4); GT vs. TT; (GG+GT) vs. TT]. In the sensitivity analyses based on the source of control, we found that the TIM-3 +4259T/G polymorphism was associated with an increased cancer risk in the hospital population [T vs. G; (GG+GT) vs.TT]. The three forests of the association between TIM-3 polymorphism (−1516G/T, −574G/T, +4259T/G) and cancer risk (the allelic model) were in Figures 2-4.
Full table
Full table
Full table
Full table
TIM-3 haplotypes
Four haplotypes were observed (GGT, TGT, GGG, and GTT) in four studies of 2,998 cases and 3,538 control subjects. Among them, the GGG haplotype increased the odds of cancer risk by 2.614-fold (OR 2.614; 95% CI: 1.756–3.893) compared with the GGT haplotype (Table 6).
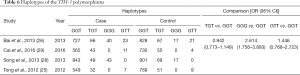
Full table
Meta-regression, sensitivity and publication bias analyses
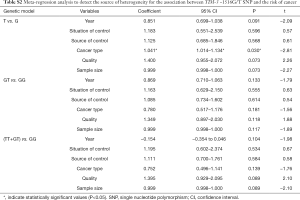
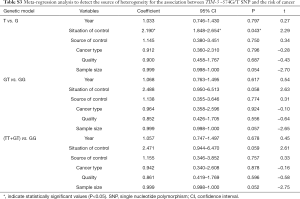
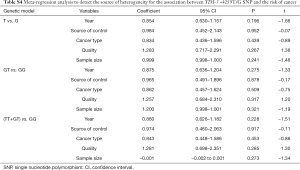
Meta-regression analysis was performed, mainly from six aspects: public year, situation of control group, source of control population, cancer types, quality of NOS scores, sample size, to identify the source of heterogeneity (Tables S2-S4). We found that the heterogeneity might be attributable to source of control population and cancer types (Tables S2,S3), and performed stratification analyses based on these outcomes.
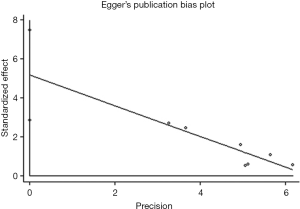
We performed a sensitivity analysis and found that there no single studies altered the pooled OR qualitatively, which indicated the stability of this meta-analysis. We then performed Egger’s test and found that those combined analysis had a publication bias under the T vs. G (Figure 5), GT vs. GG, (TT+GT) vs. GG model for the TIM-3 −1516G/T polymorphism (P=0.002, P=0.012 and P=0.017, respectively).
Discussion
In the current study, we meta-analyzed the association between TIM-3 polymorphisms and the risk of developing various human cancers in individual studies with the NOS of more than 6 scores, indicating its high quality. To our best of our knowledge, the pooled results demonstrated that the TIM-3 −1516G/T, −882C/T, −574G/T, and +4259T/G polymorphisms were associated with the susceptibility of various human cancers (i.e., gastric cancer, renal cell carcinoma, pancreatic cancer, non-small cell lung cancer, non-Hodgkin lymphomas, hepatocellular carcinoma, and thymoma), while the subgroup analyses of cancer type showed that only the TIM-3 −1516G/T polymorphism was associated with an increased cancer risk in the digestive system, whereas only the TIM-3 −574G/T polymorphism was associated with an increased risk in developing non-digestive system cancers. However, the TIM-3 +4259T/G polymorphism was not associated with any increased risk in developing both digestive system cancer and other cancers. Moreover, in the subgroup analyses of the control source, we found that four TIM-3 polymorphisms were all associated with an increased cancer risk in the hospital population in the allelic model of analysis. In terms of haplotypes, four (GGT, TGT, GGG, and GTT) occurred in TIM-3, among which the GGG haplotype was associated with an increase in the OR of cancer risk by 2.614-fold (OR 2.614; 95% CI: 1.756–3.893) compared with that of the GGT haplotype. Our current study clearly demonstrated that different TIM-3 polymorphisms contributed to human cancer risk, and further studies will disclose their potential effects on TIM-3 expression and functions in human cells.
Notably, the human TIM-3 gene contains 23,000 base pairs of DNA with 7 exons, while the TIM-3 protein was characterized by an N-terminal Ig domain of the V subset, followed by a mucin-like domain, single transmembrane domain, and a cytoplasmic tail of variable length. Different TIM-3 polymorphisms could affect TIM-3 expression and impact the protein functions, e.g., the TIM-3 +4259T/G polymorphism was reported to affect exons 3 and the mucin-like domain of the protein (10). Thus, the TIM-3 polymorphisms were associated with cancer susceptibility in human beings.
Gastric cancer is the fourth most common cancer and the second leading cause of cancer-related death in the world (30). A number of risk factors contribute to gastric cancer development, among which Helicobacter pylori infection is important (31). Chronic inflammation of the gastric mucosa induced by H. pylori could gradually progress from inflammation and premalignant changes to a suitable microenvironment for tumor initiation and establishment (32). A previous study demonstrated that TIM-3 expression was markedly increased in lymphocytes in mice infected with H. pylori and that a change in Th1 cytokine had a similar tendency as that of TIM-3 expression, while the entire bacteria and the component of H. pylori were able to promote Th1 response (33). This animal model clearly supported that infection-induced TIM-3 expression altered the host immune responses and susceptibility of gastric cancer in humans. However, the underlying mechanistic link between TIM-3 polymorphisms and the risk in developing other cancers, such as non-small cell lung cancer, is unknown; however, immune responses and overall linking to tumor immunology and aberrant immune responses affect tumor development and progression. Thus, the study of TIM-3 in immune checkpoints and modulation could help researchers better understand tumor immunology and therapy (7).
Notably, a previous meta-analysis also showed an association between TIM-3 polymorphism and cancer risk (10). However, there are many differences between the current meta-analysis and the previous one (10), e.g., the current study analyzed four TIM-3 polymorphisms (−1516G/T, −574G/T, −882C/T, and +4259T/G), whereas the previous study only analyzed three; the current study also included an analysis of four TIM-3 haplotypes (GGT, TGT, GGG, and GTT). Moreover, we added a Chinese study (9) to our subgroup analyses. However, the further study of TIM-3 polymorphisms should be extended to other human cancers to accumulate sufficient cases and controls for individual cancer sites because different cancers carry different risk factors and molecular mechanisms of carcinogenesis. For example, exposure to various environmental factors, such as tobacco smoking, could lead to lung cancer development, while an increase in red meat intake could associate with colorectal and breast cancers. How these factors coordinate with TIM-3 polymorphisms to increase the risk of these cancers requires further study. In the current meta-analysis, we did not include these environmental factors for data analysis because (I) the original studies may not have these data and (II) the inclusion of these factors could cause heterogeneity and inclusive data.
In conclusion, the current meta-analysis demonstrated that these TIM-3 polymorphisms were associated with an increased risk in the development of human cancers. However, the results were obtained through sampling statics and statistical differences, which is not the same as a clinical difference; thus, the result can be only used for clinical reference and not for clinical diagnosis or the prediction of cancer development or risk.
Acknowledgments
The authors would like to thank Medjaden Bioscience Limited.
Funding: This research was supported by the Natural Science Foundation of Hubei Provincial Department of Education (No. Q20182105), Chen Xiao-Ping Foundation for the Development of Science and Technology of Hubei Provincial (No. CXPJJH11800001-2018333), Natural Science Foundation of Hubei Province of China (No. 2016CFB530) and Faculty Development Foundation of Hubei University of Medicine (No. 2014QDJZR01), and National Students’ platform for innovation and entrepreneurship training program (No. 201810929005, 201810929009, 201810929068, and 201813249010).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Anderson AC. Tim-3: an emerging target in the cancer immunotherapy landscape. Cancer Immunol Res 2014;2:393-8. [Crossref] [PubMed]
- Gao X, Zhu Y, Li G, et al. TIM-3 expression characterizes regulatory T cells in tumor tissues and is associated with lung cancer progression. PLoS One 2012;7:e30676. [Crossref] [PubMed]
- Sakuishi K, Jayaraman P, Behar SM, et al. Emerging Tim-3 functions in antimicrobial and tumor immunity. Trends Immunol 2011;32:345-9. [Crossref] [PubMed]
- Zhu C, Anderson AC, Kuchroo VK. TIM-3 and its regulatory role in immune responses. Curr Top Microbiol Immunol 2011;350:1-15. [PubMed]
- Freeman GJ, Casasnovas JM, Umetsu DT, et al. TIM genes: a family of cell surface phosphatidylserine receptors that regulate innate and adaptive immunity. Immunol Rev 2010;235:172-89. [Crossref] [PubMed]
- Zhu C, Anderson AC, Schubart A, et al. The Tim-3 ligand galectin-9 negatively regulates T helper type 1 immunity. Nat Immunol 2005;6:1245-52. [Crossref] [PubMed]
- Ngiow SF, Teng MW, Smyth MJ. Prospects for TIM3-Targeted Antitumor Immunotherapy. Cancer Res 2011;71:6567-71. [Crossref] [PubMed]
- Zhang Y, Cai P, Liang T, et al. TIM-3 is a potential prognostic marker for patients with solid tumors: A systematic review and meta-analysis. Oncotarget 2017;8:31705-13. [PubMed]
- Du W, Yang M, Turner A, et al. TIM-3 as a Target for Cancer Immunotherapy and Mechanisms of Action. Int J Mol Sci 2017. [Crossref] [PubMed]
- Lee J, Phong B, Egloff AM, et al. TIM polymorphisms--genetics and function. Genes Immun 2011;12:595-604. [Crossref] [PubMed]
- Wang Z, Liu X, Wang X, et al. Polymorphisms in TIM-3 and breast cancer susceptibility in Chinese women: A case-control study. Oncotarget 2016;7:43703-12. [PubMed]
- Gao X, Yang J, He Y, et al. Quantitative assessment of TIM-3 polymorphisms and cancer risk in Chinese Han population. Oncotarget 2016;7:35768-75. [PubMed]
- Zhu ST, Cao BW, Xu CQ, et al. The Correlation between the TIM-3 Gene Promoter Polymorphisms and the Risk of Gastric Cancer. Journal of Capital Medical University 2010;31:299-303.
- Xu G, Zheng K, Lu X, et al. Association between polymorphisms in the promoter region of T cell immunoglobulin and mucin domain-3 and myasthenia gravis-associated thymoma. Oncol Lett 2015;9:1470-4. [Crossref] [PubMed]
- Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603-5. [Crossref] [PubMed]
- Haber M. Exact significance levels of goodness-of-fit tests for the Hardy-Weinberg equilibrium. Hum Hered 1981;31:161-6. [Crossref] [PubMed]
- Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med 1997;127:820-6. [Crossref] [PubMed]
- Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539-58. [Crossref] [PubMed]
- Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959;22:719-48. [PubMed]
- DerSimonian R, Laird N. Meta-analysis in clinical trials revisited. Contemp Clin Trials 2015;45:139-45. [Crossref] [PubMed]
- Thakkinstian A, McElduff P, D'Este C, et al. A method for meta-analysis of molecular association studies. Stat Med 2005;24:1291-306. [Crossref] [PubMed]
- Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994;50:1088-101. [Crossref] [PubMed]
- Egger M, Davey Smith G, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629-34. [Crossref] [PubMed]
- Cao B, Zhu L, Zhu S, et al. Genetic variations and haplotypes in TIM-3 gene and the risk of gastric cancer. Cancer Immunol Immunother 2010;59:1851-7. [Crossref] [PubMed]
- Tong D, Zhou Y, Chen W, et al. T cell immunoglobulin- and mucin-domain-containing molecule 3 gene polymorphisms and susceptibility to pancreatic cancer. Mol Biol Rep 2012;39:9941-6. [Crossref] [PubMed]
- Bai J, Li X, Tong D, et al. T-cell immunoglobulin- and mucin-domain-containing molecule 3 gene polymorphisms and prognosis of non-small-cell lung cancer. Tumour Biol 2013;34:805-9. [Crossref] [PubMed]
- Li Z, Li N, Zhu Q, et al. Genetic variations of PD1 and TIM3 are differentially and interactively associated with the development of cirrhosis and HCC in patients with chronic HBV infection. Infect Genet Evol 2013;14:240-6. [Crossref] [PubMed]
- Song H, Ma S, Cha Z, et al. T-cell immunoglobulin- and mucin-domain-containing molecule 3 genetic variants and HIV+ non-Hodgkin lymphomas. Inflammation 2013;36:793-9. [Crossref] [PubMed]
- Cai C, Xu YF, Wu ZJ, et al. Tim-3 expression represents dysfunctional tumor infiltrating T cells in renal cell carcinoma. World J Urol 2016;34:561-7. [Crossref] [PubMed]
- Van Cutsem E, Sagaert X, Topal B, et al. Gastric cancer. Lancet 2016;388:2654-64. [Crossref] [PubMed]
- Li L, Ying XJ, Sun TT, et al. Overview of methodological quality of systematic reviews about gastric cancer risk and protective factors. Asian Pac J Cancer Prev 2012;13:2069-79. [Crossref] [PubMed]
- Moss SF, Blaser MJ. Mechanisms of disease: Inflammation and the origins of cancer. Nat Clin Pract Oncol 2005;2:90-7; quiz 1 p following 113.
- Hu S, Xie Y, Zhou N, et al. Expression of T-cell immunoglobulin- and mucin-domain-containing molecules-1 and -3 (Tim-1 and Tim-3) in Helicobacter pylori infection. Helicobacter 2011;16:373-81. [Crossref] [PubMed]