
Rapid and sensitive detection of Shigella flexneri using fluorescent microspheres as label for immunochromatographic test strip
Introduction
Shigella can cause an acute bloody diarrhea in humans which is becoming an increasing public health burden due to development of multiple antimicrobial resistance frequently resulting in treatment failure (1,2). It has been estimated that 165 million cases of shigellosis occur annually worldwide, resulting in 1.1 million deaths (3-5).
At present, the traditional detection technologies of microbiological cultivation and drug resistance are laborious, time-consuming and insensitive, it generally takes 2–3 days to complete the steps of enrichment and confirmation. Molecular detection methods are characterized by rapid identification, high sensitivity and specificity compared with the culture methods. Two approaches have emerged: polymerase chain reaction (PCR) (6,7) and immunochromatographic techniques (8-15). The traditional PCR assay based on agarose gel electrophoresis (PCR-GE) and real-time fluorescent quantitative PCR (RTFQ-PCR) methods are now widely used in the pathogenic microorganism detection field. However, PCR-GE can only perform qualitative test and the following nucleic acid dyes in agarose gel electrophoresis step are also toxic. The equipment of RTFQ-PCR is expensive, which is adverse to the extensive use in the clinical practice.
Immunochromatographic technology was first introduced in the 1980s. Design of immunochromatographic test strip (ICTS) can be currently used for pathogens’ nucleic acid detection, and is called “nucleic acid lateral flow immunoassay” (NALFIA) (8,16-19). Several recent reports have described the detection of PCR amplicon in flow with functionalized nanoparticles on the strip (16-18,20-24). To our knowledge, immunochromatographic technology is most used for qualitative experiments (9-15,25-27) and their sensitivity is worrying.
In this study, PCR-ICTS method, as a novel rapid, quantifiable, sensitive and specific technique, which is conducted by introducing magnetic beads purification, biotin-streptavidin system and fluorescent microspheres (FMs) for the detection of clinical samples of Shigella flexneri (S. flexneri). The magnetic beads enable the isolation or extraction of target molecule or substance due to the good biocompatibility and adequate functional groups for chemical fixation which have been applied to immunoassay (28,29). Silicon coated magnetic nanoparticles (Si-MNPs) we made can be used for specific bioaffinity capture of molecules. Moreover, FMs have good thermal stability, dispersibility, biocompatibility, high fluorescence stability, surface modification, narrow particle size and high luminous efficiency (30-33), and FMs are rarely employed for the immunochromatography methods. In addition, the genus-specific genes of ipaH (invasion plasmid antigen H) (34) and drug resistant gene of CTX-M-9 that encoding CTX-M ESBLs (extended spectrum β-lactamase) (35) from Shigella were selected to investigate the potential of the new immunochromatographic technology in early clinical detection as well as to compare the currently used classical microbiological methods (API Coryne System), PCR-GE and the RTFQ-PCR method.
Methods
Microbial cultivation conditions and identification
The strains used in this study were listed in Table 1. Sixty-three Shigella strains of the clinical samples were provided by Jiangsu Provincial Center for Disease Control and Prevention (Xuzhou, China). Additional, non-Shigella strains were selected from Affiliated Hospital of Xuzhou Medical University (Xuzhou, China) to assess the specificity of PCR-ICTS. The standard strain used in this study was S. flexneri (ATCC 12022) which was obtained from Bena Culture Collection (Beijing, China). Strains identification was performed by API Coryne System (BioMerieux, France) and drug sensitivity experiments were performed by VITEK-2 Compact automatic microbial analysis system (Merieux, France) and DNA sequencing.

Full table
DNA extraction
The DNA extractions from all samples were performed by TIANamp Bacterial Genomic DNA kit (Tiangen, Beijing, China), according to the manufacturer’s protocols. The quality of DNA was evaluated by measuring the ratio of A260/A280 on an ultraviolet spectrophotometer (Merinton SMA 1000, Beijing, China) and detecting integrity with agarose electrophoresis. The genomic DNA (gDNA) of the strains above was stored at −80 °C until they were thawed on ice immediately prior to analysis.
PCR applications and agarose gel electrophoresis
Upstream primer and downstream primer of two marker genes (ipaH, CTX-M-9) (34,35) from Shigella were synthesized and labelled on 5'-end with biotin and digoxigenin, respectively, by Sangon Biotech Co., Ltd. (Shanghai, China). The gene CTX-M-9 has been identified with a high positive rate detected by VITEK-2 Compact and DNA sequencing in our previous literature (36). The primers used to amplify the target sequences of S. flexneri and their sequences were listed in Table 2. The PCR reaction mixture contained 10 µL of Green Taq mix (Vazyme, Nanjing, China), 0.5 µL of forward primer (10 µmol/L), 0.5 µL of reverse primer (10 µmol/L), 1 µL of sample DNA and sterile deionized water (Tiangen, Beijing, China) to a final reaction volume of 20 µL. Direct PCRs were performed using a Bio-Rad PCR System (Shanghai, China) under the following conditions: 94 °C for 5 min, 30 cycles of 94 °C for 50 s, 57.5 °C for 50 s (ipaH) or 60 °C for 30 s (CTX-M-9) and 72 °C for 1 min, and a final extension at 72 °C for 5 min. The amplification process was completed in approximately 2 h. The PCR products were divided into two parts, one for electrophoresis analysis and the other for PCR-ICTS detection. In PCR-GE analysis, the products of PCR were analyzed by electrophoresis in 1.5% (w/v) agarose gel with gelred nucleic acid dye and the DNA bands were visualized by gel imaging system (Bioshine GelX 1850, Shanghai, China) and compared to DL1000 Marker (TaKaRa, Dalian, China) to verify the size of amplicons.

Full table
PCR-ICTS detection
Purification of PCR products
PCR products of Shigella above were carried out for purification as follows: 5 µL PCR products added 10 µL of Si-MNPs and the mixture was incubated at room temperature (RT) with gentle mixing for 5 min so that the DNA fragments could be completely adsorbed. Then SiMNPs-DNA complex was immobilized by a magnet for 2 min. After washing 2 times with ethanol solution (85%), the Si-MNPs were resuspended by 15 µL of sterile deionized water, and then DNA was released from the Si-MNPs. The sterile deionized water suspended contained PCR product was placed at RT for 5 min to fully eluted and the purified PCR product could be used for immunochromatographic detection or stored at −20 °C. The overall process is shown in Figure 1.
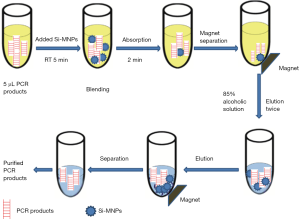
Detection of purification of PCR products by ICTS
The reaction mixture containing 15 µL of purified PCR products was added onto the sample pad of ICTS we made (Nanoeast, Nanjing, China) and then 85 µL of PBS (pH 7.4) was added, the reaction mixture was localized to the specific positions which were designated as the capture test lines on the strip, and 100 µL PBS solution (pH 7.4) was used as the blank control. After 5 min the results were observed by the test strip reader of Nanoeasy 1700 which we made. To evaluate the repeatability, all samples were detected triplicates.
Detection of sensitivity and specificity of ICTS
The sensitivity of ICTS (ipaH) was evaluated by gDNA of S. flexneri strain at different concentration (2.5×100–2.5×10−8 ng/µL) prepared by dilutions with free deionized water and compared with the results of gel electrophoresis (Figure 2). Standard DNA is diluted from 25 to 2.5×10−8 ng/µL for PCR amplification to evaluate the detection limit of PCR-GE and PCR-ICTS. Free deionized water was used as negative control and the tests were repeated three times at each concentration. And the last dilution that yielded a positive result was regarded as the detection limit of the test strip. With the test strip of reader Nanoeasy 1700, results can be quantified and shared in real time via the internet. The threshold value of fluorescence signal value was defined as 500 (Cutoff value) on the test line. If the signal value was greater than 500, it indicated that the result was positive, and vice versa, a negative outcome would be indicated by the signal value less than 500 at the test regions.
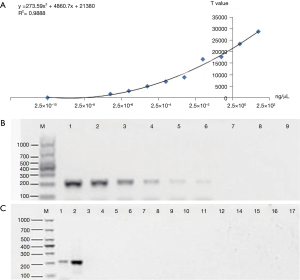
ipaH was used to detect the specificity of Shigella (Figure 2C), samples of S. flexneri and the other pathogenic microorganism (11 gram-negative bacteria, 4 gram-positive bacteria and 1 Candida albicans) were used to assess the specificity of the test strips. 20 cases of S. flexneri were selected from 2012 to 2015. The non-Shigella strains used in this study were listed in Table 1. The results of ICTS were compared with PCR-GE and API Coryne System to determine the specificity and accuracy of ICTS.
The quantitative detection by RTFQ-PCR
RTFQ-PCR reaction was carried out on a cobas®z 480 Automatic Fluorescence Quantitative PCR Analyzer (ABI, America) using a SYBR Green PCR kit (Vazyme, Nanjing, China). Each 20 µL reaction mixture contained 10 µL AceQ® qPCR SYBR® Green Master Mix, 0.4 µL primer-F (10 µmol/L), 0.4 µL primer-R (10 µmol/L), 0.4 µL of 50× ROX Reference Dye1, 1 µL of DNA templates, and sterile deionized water was used to make the volume up to 20 µL. Cycling conditions were as following: an initial denaturation at 95 °C for 5 min, 30 cycles of 95 °C, 10 s, 60 °C, 50 s, 72 °C, 1 min and a final elongation 72 °C for 5 min. The sensitivity of RTFQ-PCR (ipaH) was evaluated by gDNA of S. flexneri strain at different copy number (2.0×108–2.0×104 copies/µL). All runs were repeated in triplicate including a negative control and positive control. Eleven cases of S. flexneri were selected from 2012 to 2015 to detect genus-specific genes (ipaH) of S. flexneri by PCR-ICTS and RTFQ-PCR.
Results
The principle of ICTS
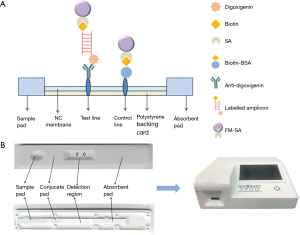
The ICTS consisted of six parts: the sample pad, the nitrocellulose membrane (NC), the conjugate pad, the absorption pad, the polystyrene backing card as well as a detection region with test and control lines. gDNA isolated from the analyzed pathogenic bacteria was amplified using PCR with two tagged primers specific to S. flexneri. The amplified PCR product is labeled with biotin at one end, combined with FM-labeled streptavidin, and the other end is labeled with digoxigenin, which binds to the anti-digoxigenin antibody on the test line, thereby fluorescence detection system receives fluorescence signal. The more FMs are combined, the stronger fluorescent signal can be detected, with the test strip of reader Nanoeasy 1700, results can be quantified and shared in real time via the internet. However, excess streptavidin-coated FM uncomplexed with biotin-labeled PCR products conjugated with the biotin at the control line (Figure 3A). The response is directly proportional to the amount of analyte and quantitative data may be obtained by recording the optical responses with a portable strip reader. A detailed schematic of the ICTS was given in Figure 3A and the photographs of the complete ICTS in its polystyrene casing and the internal structure were shown in Figure 3B.
The positive result appeared with two data at the test and control line positions on the strip, whereas the negative result with very low value (value <500) at the test line position. The negative control, for which PBS was used instead of PCR products, did not result in any value on the test lines. Thus, we confirmed that the ICTS can be utilized for detection of Shigella spp. No false-positive or false-negative results were ever seen with the ICTS, and the results were comparable to PCR-GE and RTFQ-PCR (Table 1). Characteristics of the common methods for detecting Shigella were shown in Table 3.

Full table
Sensitivity of PCR-ICTS
To determine the sensitivity of ipaH detection by PCR-GE and PCR-ICTS, the standard DNA stock (25 ng/µL) was serially diluted tenfold (Figure 2). Each gDNA (1 µL) was used as a template for PCR where the respective dilutions were subjected to thermal cycling using the primer pair of biotin-labeled ipaH-F and digoxigenin-labeled ipaH-R, which amplified a 204-bp fragment from the ipaH gene of S. flexneri. In PCR-ICTS, standard DNA dilutions gave a fluorescence signal values ranging from 32.85 to 28,733, and a linear relationship (y=273.59x2+4860.7x+21380) was observed with ipaH titers decreasing from 2.5×100 to 2.5×10−8 ng/µL. The correlation analysis revealed that the Pearson correlation coefficient (Pearson’s r) between DNA concentration and ICTS results was 0.9888 (R2=0.9888) (Figure 2A). The results showed that the sensitivity of PCR-ICTS for the detection of ipaH was 2.5×10−7 ng/µL, which was 100 times more sensitive than PCR-GE assay (Figure 2).
Comparison of ICTS and RTFQ-PCR
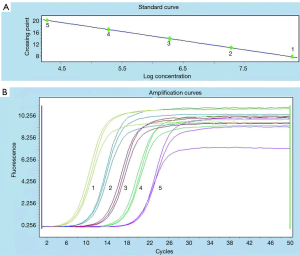
According to Figure 4A, there was a good linear relationship between the logarithm of DNA concentration and the cycle threshold (Ct) value of RTFQ-PCR. The linear equation was y=−3.194x+33.95, with a R2 value of more than 0.99. The minimum detection limit for plasmids containing target genes was 2 copies/µL, which was converted to a DNA concentration with a sensitivity of 3.2×10−7 ng/µL. From Figure 4B, it could be judged that the detection results of different DNA concentrations of S. flexneri had very good repeatability. At the same time, combined with the results of Figures 2C,4, the above results indicated that the selected ipaH gene and specific primers could separate Shigella from other bacteria, and had very good repeatability and sensitivity. It can be seen that the sensitivity of ICTS is basically the same as the sensitivity of RTFQ-PCR, but the cost of PCR-ICTS is low (equipment costs less than 30,000 RMB), and the result of ICTS is easier to interpret.
Specificity of PCR-ICTS
The specificity of the PCR-ICTS was determined in the presence of fifteen non-Shigella samples. To validate the specificity of the ICTS, the purified PCR product of ipaH was analyzed using the ICTS and the agarose gel electrophoresis, separately. Table 1 demonstrated interpretation of ICTS and electrophoresis results in the recognition of Shigella and non-Shigella from different microbial samples. The electrophoretic images showed that all the amplified products of ipaH had the correct bands of 204 bp, and non-Shigella had no bands (Figure 2C). The results were consistent with the ICTS results, which indicated that the ICTS exhibits a high degree of specificity.
The results of before and after purification of PCR products
To obtain the clarification of PCR product, Si-MNPs were used to purify the amplicons to remove the false positive results. As shown in Table 4, the value of test line in the dilution group (blank control) showed a low background signal value (i.e., low fluorescence intensity), indicating that the test strip itself did not produce false positives. In the negative control group, the fluorescence intensity of the signal before purification of the negative sample was 1,628, indicating that non-target band was amplified to cause a false positive result, the signal intensity after purification was reduced to 180, indicating that most of the non-target bands (including primer dimers) were removed during the purification process and ICTS could be utilized for the detection of Shigella. There was no significant difference in the signal values between the positive samples H1, H2 and H3 before and after the purification, and the signal values were strong, indicating that the purification process can filter the target bands very strongly. These results were consistent with the results of agarose gel electrophoresis. Thus, the test strip had good repeatability and strong screening ability, which can remove false positive results.

Full table
2nd application: detection of drug resistance genes of S. flexneri
Sixty-three Shigella strains were selected for the detection of drug resistance genes of CTX-M-9. All Shigella strains of the clinical samples were identified by API Coryne System and their drug resistance had also been verified by VITEK-2 Compact and DNA sequencing. The CTX-M-9 gene from Shigella was amplified by PCR using primers (Table 2). The PCR products obtained were detected by agarose gel electrophoresis and ICTS, respectively. The results are show in Table 5, the results of PCR-ICTS were in accordance with the traditional VITEK-2 Compact combine with DNA sequencing. However, the target band of three PCR products obtained by agarose gel electrophoresis was weak, but they still showed higher fluorescence intensity when tested with ICTS (Table 5). This result shows that the ICTS has good sensitivity and strong screening ability which can remove false negative results.

Full table
Discussion
The drug resistant genes of CTX-M group have become the most common type of ESBL in Asia and Latin America (35-39), the presence of CTX-M-9 in all of the resistance to β-lactams mediated by CTX-M enzymes was 49.2%, of which is a common CTX-M ESBLs gene (35,36,38,40,41). In this study, 30 (47.6%) samples were positive by using PCR-ICTS and traditional VITEK-2 Compact combine with DNA sequencing, the target band of three PCR products obtained by agarose gel electrophoresis were weak, but they still showed higher fluorescence intensity when tested with ICTS. This result shows that the ICTS has good sensitivity and strong screening ability which can remove false negative results. In addition, ICTS can simultaneously detect Shigella spp. and its drug resistance gene of pathogenic microorganisms and provide effective method for clinical accurate detection of pathogenic microorganism infections and drug resistance, and provide reference for clinician using drugs. This paper is the first to use ICTS to detect the third-generation cephalosporin resistance gene of pathogenic microorganisms. The minimum detectable DNA concentration of the ICTS was 2.5×10−7 ng/µL after amplificatin under optimized condition. However, the conventional PCR assay based on gel electrophoresis only reached 2.5×10−5 ng/µL, which further demonstrated the PCR-ICTS had good sensitivity than PCR-GE.
As previous literatures reported, the immunochromatographic method can only detect one specific bacterium based on monoclonal or polyclonal antibodies (42-47), and most test strips are only used for qualitative experiments (9-15,25-27). In this study, FMs were used to bind streptavidin to capture biotin-labeled PCR product. Compared with colloidal gold, FMs have higher signal values, wider linear range, better interbatch repeatability and it is also suitable for the quantitative detection (48). Moreover, the highly specific affinity and multistage amplification effect between biotin and FM-SA could be used at the test lines to improve the sensitivity of immune binding and tracer analysis. In addition, some ICTS-based methods need large sample and require special equipments as well as complex analysis process. For example, Xu et al. had presented a novel ICTS using FMs labeled with polyclonal antibodies for the detection of Campylobacter jejuni (C. jejuni), which can only qualitatively detect C. jejuni (43). The ICTS developed by Zhang Fan based on lanthanide chelate-loaded silica nanoparticles had been used for the rapid detection of Pantoea stewartii subsp. stewartii (Pss), but the more samples were required (45). Blažková et al. developed a ICTS detection method, which requires special equipments and complex analysis process (with a total analysis time of 16 hours) (49). The PCR-ICTS can be used for rapid quantitative detection clinical pathogens based on the PCR products, which can greatly increase the detection range of pathogenic bacteria. The specially labeled PCR amplification products were purified by magnetic beads and put into the prepared FMs-ICTS for quantitative detection. The sensitivity of the detection scheme reached 25 fg/µL DNA samples through the exploration and optimization of sensitivity, specificity and other parameters. Compared with the traditional clinical method, this method has a fast detection speed, reduces the detection workload and reduces the probability of bacterial contamination, whereas the traditional microbiological method usually require 2–3 days to obtain a definitive result, which are time consuming and labor intensive (49,50).
However, ICTS provides following advantages over gel electrophoresis in detection of PCR product: (I) Compared with electrophoresis (90 min), it takes only 30 min (25 min of purification and 5 min of ICTS) to finish the detection; (II) it is capable of quantitative detection; (III) no dangerous compounds is used while nucleic acid dye is used in electrophoresis. Compared with conventional PCR, this method can be used for quantitative detection of target genes. Compared with the existing shigella immune chromatography technology, the developed ICTS can simultaneously detect gene- specific genes and drug resistance genes. Thus, the ICTS can instead of gel electrophoresis to detect PCR products due to its specificity, speed, simplicity and low-cost. Compared with the RTFQ-PCR method, PCR-ICTS provides several advantages such as low detection cost, no technical expertise in performing the test and interpreting the results as well as the compact size (the same size as A4 paper) of the strip reader. Moreover, SYBR green I dye used in RTFQ-PCR method can combine any double-stranded DNA, which might produce false positive results. However, the products in PCR-ICTS must be purified before using the ICTS, so the PCR-ICTS could reduce false positive results. Hence this plan is more advantageous to clinical and medical equipment backward area of promotion, the preparation can be realized rapid detection kit of shigella and their drug resistance gene quantitative test of low cost and high sensitivity and precision.
The objective of this study was to eliminate the use of gel electrophoresis in PCR product analysis and replaced it with ICTS which could offer a more rapid, quantitative and cost-effective method. Moreover, the sensitivity of the ICTS was consistent with that of RTFQ-PCR, but the cost was much cheaper than that of RTFQ-PCR, which was more suitable for the detection of clinical samples, especially for the intestinal pathogens in poor working conditions of hospital, which have few money to buy expensive medical equipments. Therefore, the developed PCR-ICTS can be used as the ideal tool for the quantitative and accurate detection of important intestinal pathogens such as Shigella to immediately confirm the patients who contact with the pathogen due to its sensitivity, specificity, simplicity, low-cost and better detectability.
In this paper, ICTS can detect amplification products of a single gene at a time, then we will design multi-channel immunochromatography to rapidly identify more pathogens, hoping to help clinicians diagnose early. In addition, we also carried out rolling circle amplification experiments, applied to immunochromatography, which can further improve the overall sensitivity, provide an effective promotion program for clinical accurate detection, diagnosis and treatment of intestinal important pathogen infection.
Conclusions
This work described a FM-based ICTS method to realizing the detection of S. flexneri as low as 2.5×10−7 ng/µL in 3 h. The results demonstrated that the novel PCR-ICTS method can instead of PCR-GE and RTFQ-PCR to detect Shigella due to its speed, non-poisonous, simplicity and low-cost. In particular, this PCR-ICTS method displayed high specificity and sensitivity without the requirement of very specific and expensive equipment as well as the complicated techniques, which is convenient for the rapid detection of enteric pathogens and has a potential to be a useful tool for the rapid and easy-to-use detection of clinical samples.
Acknowledgments
Funding: The work was supported by grants from the National Natural Science Foundation of China (81702103), Jiangsu Provincial Natural Science Foundation (BK20170252), Projects for Jiangsu Provincial Young Medical Talents (QNRC2016780), General Program of the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (16KJD320005), and Xuzhou Science and Technology Planning Project (KC16SY157).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Khan WA, Griffiths JK, Bennish ML. Gastrointestinal and extra-intestinal manifestations of childhood shigellosis in a region where all four species of Shigella are endemic. PLoS One 2013;8:e64097. [Crossref] [PubMed]
- Li Y, Cao B, Liu B, et al. Molecular detection of all 34 distinct O-antigen forms of Shigella. J Med Microbiol 2009;58:69-81. [Crossref] [PubMed]
- Agtini MD, Soeharno R, Lesmana M, et al. The burden of diarrhoea, shigellosis, and cholera in Jakarta, Indonesia. BMC Infect Dis 2005;5:89. [Crossref] [PubMed]
- Wang XY, Du L, Von Seidlein L, et al. Occurrence of shigellosis in the young and elderly in rural China: results of a 12-month population-based surveillance study. Am J Trop Med Hyg 2005;73:416-22. [Crossref] [PubMed]
- Gupta A, Polyak CS, Bishop RD, et al. Laboratory confirmed shigellosis in the United States, 1989-2002: Epidemiologic trends and patterns. Clin Infect Dis 2004;38:1372-7. [Crossref] [PubMed]
- von Seidlein L, Kim DR, Ali M, et al. A Multicentre Study of Shigella Diarrhoea in Six Asian Countries: disease burden, clinical manifestations, and microbiology. PLoS Med 2006;3:e353. [Crossref] [PubMed]
- Vu DT, Sethabutr O, Von Seidlein L, et al. Detection of Shigella by a PCR assay targeting the ipaH gene suggests increased prevalence of shigellosis in Nha Trang, Vietnam. J Clin Microbiol 2004;42:2031-5. [Crossref] [PubMed]
- Posthuma-Trumpie GA, Korf J, van Amerongen A. Lateral flow (immuno) assay: its strengths, weaknesses, opportunities and threats. A literature survey. Anal Bioanal Chem 2009;393:569-82. [Crossref] [PubMed]
- Taneja N, Nato F, Dartevelle S, et al. Dipstick Test for Rapid Diagnosis of Shigella dysenteriae 1 in Bacterial Cultures and Its Potential Use on Stool Samples. PLoS One 2011;6:e24830. [Crossref] [PubMed]
- Nato F, Phalipon A, Nguyen TL, et al. Dipstick for rapid diagnosis of Shigella flexneri 2a in stool. PLoS One 2007;2:e361. [Crossref] [PubMed]
- Haddar C, Begaud E, Maslin J, et al. Point-of-care tests for the rapid diagnosis of shigellosis. Bull Soc Pathol Exot 2017;110:1-8. [Crossref] [PubMed]
- Chau ML, Hartantyo SH, Yap M, et al. Diarrheagenic pathogens in adults attending a hospital in Singapore. BMC Infect Dis 2016;16:32. [Crossref] [PubMed]
- Varela G, Batthyány L, Bianco MN, et al. Enteropathogens associated with acute diarrhea in children from households with high socioeconomic level in Uruguay. Int J Microbiol 2015;2015:592953. [PubMed]
- Sire JM, Garin B, Chartier L, et al. Community-acquired infectious diarrhoea in children under 5 years of age in Dakar, Senegal. Paediatr Int Child Health 2013;33:139-44. [Crossref] [PubMed]
- Bonkoungou IJ, Haukka K, Österblad M, et al. Bacterial and viral etiology of childhood diarrhea in Ouagadougou, Burkina Faso. BMC Pediatr 2013;13:36. [Crossref] [PubMed]
- Kozwich D, Johansen KA, Landau K, et al. Development of a Novel, Rapid Integrated Cryptosporidium parvum Detection Assay. Appl Environ Microbiol 2000;66:2711-7. [Crossref] [PubMed]
- Fong WK, Modrusan Z, McNevin JP, et al. Rapid solid-phase immunoassay for detection of methicillin-resistant Staphylococcus aureus using cycling probe technology. J Clin Microbiol 2000;38:2525-9. [PubMed]
- Soo PC, Horng YT, Hsueh PR, et al. Direct and simultaneous identification of Mycobacterium tuberculosis complex (MTBC) and Mycobacterium tuberculosis (MTB) by rapid multiplex nested PCR-ICT assay. J Microbiol Methods 2006;66:440-8. [Crossref] [PubMed]
- Edwards KA, Baeumner AJ. Optimization of DNA-tagged dye-encapsulating liposomes for lateral-flow assays based on sandwich hybridization. Anal Bioanal Chem 2006;386:1335-43. [Crossref] [PubMed]
- Andrade TP, Lightner DV. Development of a method for the detection of infectious myonecrosis virus by reverse-transcription loop-mediated isothermal amplification and nucleic acid lateral flow hybrid assay. J Fish Dis 2009;32:911-24. [Crossref] [PubMed]
- Blazková M, Koets M, Rauch P, et al. Development of a nucleic acid lateral flow immunoassay for simultaneous detection of Listeria spp. and Listeriamonocytogenes in food. Eur Food Res Technol 2009;229:867-74. [Crossref]
- Dineva MA, Candotti D, Fletcher-Brown F, et al. Simultaneous Visual Detection of Multiple Viral Amplicons by Dipstick Assay. J Clin Microbiol 2005;43:4015-21. [Crossref] [PubMed]
- Mao X, Ma Y, Zhang A, et al. Disposable Nucleic Acid Biosensors Based on Gold Nanoparticle Probes and Lateral Flow Strip. Anal Chem 2009;81:1660-8. [Crossref] [PubMed]
- Mens PF, van Amerongen A, Sawa P, et al. Molecular diagnosis of malaria in the field: development of a novel one-step nucleic acid lateral flow immunoassay for the detection of all 4 human Plasmodium spp. and its evaluation in Mbita, Kenya. Parasitology 2008;61:412-27.
- Pruksaphon K, Intaramat A, Ratanabanangkoon K, et al. Development and characterization of an immunochromatographic test for the rapid diagnosis of Talaromyces (Penicillium) marneffei. PLoS One 2018;13:e0195596. [Crossref] [PubMed]
- Tinky SS, Ambily R, Nair SR, et al. Utility of a rapid immunochromatographic strip test in detecting canine parvovirus infection compared with polymerase chain reaction. Vet World 2015;8:523-6. [Crossref] [PubMed]
- Siu LK, Tsai YK, Lin JC, et al. Development of a Colloidal Gold-Based Immunochromatographic Strip for Rapid Detection of Klebsiella Pneumoniae Serotypes K1 and K2. J Clin Microbiol 2016;54:3018-21. [Crossref] [PubMed]
- Chen WJ, Tsai PJ, Chen YC. Functional nanoparticle-based proteomic strategies for characterization of pathogenic bacteria. Anal Chem 2008;80:9612-21. [Crossref] [PubMed]
- Ravindranath SP, Mauer LJ, Deb-Roy C, et al. Biofunctionalized magnetic nanoparticle integrated mid-infrared pathogen sensor for food matrixes. Anal Chem 2009;81:2840-6. [Crossref] [PubMed]
- Bradley M, Ashokkumar M, Grieser F. Sonochemical production of fluorescent and phosphorescent latex particles. J Am Chem Soc 2003;125:525-9. [Crossref] [PubMed]
- Duong HD, Rhee J I. Use of CdSe/ZnS core-shell quantum dots as energy transfer donors in sensing glucose. Talanta 2007;73:899-905. [Crossref] [PubMed]
- Hirschey MD, Han YJ, Stucky GD, et al. Imaging Escherichia coli using functionalized core/shell CdSe/CdS quantum dots. J Biol Inorg Chem 2006;11:663-9. [Crossref] [PubMed]
- Liu W, Howarth M, Greytak AB, et al. Compact biocompatible quantum dots functionalized for cellular imaging. J Am Chem Soc 2008;130:1274-84. [Crossref] [PubMed]
- Li ZQ, Deng SL, Sun CM. Detection of ipaH gene of Shigella by PCR. Chinese J Clin Lab Sci 2009;27:185-6.
- Yu Y, Ji SJ, Chen YG, et al. Resistance of strains producing extended-spectrum β-lactamases and genotype distribution in China. J Infect 2007;54:53-7. [Crossref] [PubMed]
- Liu G, Qian H, Tang B, et al. Prevalence and characterization of third generation cephalosporin resistant Shigella flexneri isolates from Jiangsu of China, 2013-2015. J Glob Antimicrob Resist 2018;15:283-7. [Crossref] [PubMed]
- Hawkey PM. Prevalence and clonality of extended-spectrum-lactamases in Asia. Clin Microbiol Infect 2008;14:159-65. [Crossref] [PubMed]
- Bialvaei AZ, Kafil HS, Asgharzadeh M, et al. CTX-M extended-spectrum β-lactama-producing Klebsiella spp, Salmonella spp, Shigella spp and Escherichia coli isolates in Iranian hospitals. Braz J Microbiol 2016;47:706-11. [Crossref] [PubMed]
- Ning YZ, Wang H, Sun HL, et al. The molecular epidemiology of extended-spectrum β-lactamases (ESBL) and plasmidz-mediated AmpC β- factamases (p-AmpC) produced by the clinical strains of E. coli and K. pneumoniae. Chinese J Microbiol Immunol 2006;26:944-9.
- Kim S, Kim J, Kang Y, et al. Occurrence of extended-spectrum beta-lactamases in members of the genus Shigella in the Republic of Korea. J Clin Microbiol 2004;42:5264-9. [Crossref] [PubMed]
- Park SY, Kang CI, Wi YM, et al. Risk factors and molecular epidemiology of community-onset, multidrug resistance extended-spectrum β-lactamase- producing Escherichia coli infections. Korean J Intern Med 2017;32:146-57. [Crossref] [PubMed]
- Feng M, Kong D, Wang W, et al. Development of an Immunochromatographic Strip for Rapid Detection of Pantoea stewartii subsp. Stewartii. Sensors (Basel) 2015;15:4291-301. [Crossref] [PubMed]
- Xu D, Wu X, Li B, et al. Rapid detection of Campylobacter jejuni using fluorescent microspheres as label for immunochromatographic strip test. Food Sci Biotechnol 2013;22:585-91. [Crossref]
- Wen-de W, Min L, Ming C, et al. Development of a colloidal gold immunochromatographic strip for rapid detection of Streptococcus agalactiae in tilapia. Biosens Bioelectron 2017;91:66-9. [Crossref] [PubMed]
- Zhang F, Li J, Zou M, et al. Simultaneous Detection of Clavibacter michiganensis subsp nebraskensis and Pantoea stewartii subsp stewartii Based on Microsphere Immunoreaction. J Biomol Screen 2013;18:474-80. [Crossref] [PubMed]
- Werawatganon D, Pongsuchart M, Sereemaspun A, et al. Development of a Lateral-flow Immunochromatographic Strip Using Gold Nanoparticles for Helicobacter pylori Detection. J Med Assoc Thai 2015.Suppl 1:S85-90. [PubMed]
- Kong MM, Yang B, Gong CJ, et al. Development of immunochromatographic colloidal gold test strip for rapid detection of Haemophilus influenzae in clinical specimens. J Appl Microbiol 2017;123:287-94. [Crossref] [PubMed]
- Chen X, Xu H, Lai W, et al. A sensitive chromatographic strip test for the rapid detection of enrofloxacin in chicken muscle. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 2012;29:383-391. [PubMed]
- Blažková M, Javůrková B, Fukal L, et al. Immunochromatographic strip test for detection of genus Cronobacter. Biosens Bioelectron 2011;26:2828-34. [Crossref] [PubMed]
- Gracias KS, McKillip JL. A review of conventional detection and enumeration methods for pathogenic bacteria in food. Can J Microbiol 2004;50:883-90. [Crossref] [PubMed]


