
‘RNA circles of influence’ in Kaposi sarcoma
Circular RNAs (circRNAs) are increasingly implicated in different biological processes. A recent report by Tagawa et al., Proc Natl Acad Sci U S A [2018] investigates the circRNAs expressed following infection by Kaposi sarcoma herpesvirus (KSHV). We discuss the proposed biological functions of such circRNAs in enhancing or repressing virulence, as well as the possible utility of subsets of circRNAs in antiviral therapy.
KSHV, also known as human herpesvirus 8 (HHV-8), is a gamma herpesvirus first identified in tissues from AIDS (acquired immunodeficiency syndrome) patients with associated Kaposi sarcoma (KS) (1). KSHV was later linked to different KS forms as well as to neoplastic and lymphoproliferative diseases (2-5). KS in the United States accounts for ~4% of cases worldwide (6,7), and is among the most prominent cancers in patients with AIDS (8). Besides transcribing several protein-coding mRNAs, the KSHV genome expresses multiple noncoding RNAs (ncRNAs), including microRNAs (miRNAs) and long ncRNAs (lncRNAs). In addition, the KSHV genome can also give rise to circRNAs, as recently discovered (9-11).
CircRNAs can arise from linear precursor RNAs transcribed from coding and noncoding genes, and can contain exonic and intronic sequences. Many circRNAs are generated via backsplicing events whereby the 5' and 3' ends are covalently religated (12). Due to the lack of exposed 5' and 3' ends, circRNAs are protected by exonuclease degradation and hence are more stable than the linear precursor RNAs. Recent progress in RNA sequencing technology has revealed that circRNAs are highly abundant and are often expressed in specific tissues, drawing immense interest to understand their functions in health and disease (13-18). Some insight into circRNA function has been gained from evidence that circRNAs can interact with proteins and other RNAs (12). For instance, abundant circRNAs can bind miRNAs, acting as miRNA ‘sponges’ and inhibiting miRNA-mediated suppression of mRNAs. Similarly, the interaction of circRNAs with RNA-binding proteins (RBPs) can affect the function of RBPs upon other cellular targets (19). CircRNA interactions with other RNAs (e.g., mRNAs, lncRNAs) and with DNA have been shown or proposed to influence many cellular processes (12).
In a recent study, Tagawa and colleagues elegantly showed that upon KSHV infection, human umbilical vein endothelial cells (HUVECs) expressed many circRNAs (20). RNA-Seq analysis revealed numerous backspliced junctions from the KSHV genome, a finding that was validated by individual circRNA detection (21). By employing TREx-BCBL1 cells expressing RTA, an important modulator of the viral lytic phase, the authors discovered that viral circRNAs were more abundant in the lytic phase than the latent phase. In line with this finding, increased RTA levels in lymph node biopsies from KSHV-infected individuals correlated positively with high levels of circRNAs. As the number of tested biopsies was limited to one individual per condition, it would be important to extend these interesting observations to additional individuals in future work. KSHV-derived circRNAs kcirc55 and kcirc97 were proposed to be implicated in cell proliferation; other KSHV-encoded circRNAs were postulated to influence gene expression during apoptosis and other tumor-related processes.
Using circRNA microarrays, the authors further identified circRNAs with altered expression levels in KSHV-infected HUVECs and human B lymphocytes (MC116 cells) (20). Interestingly, several of these circRNAs harbored multiple binding sites for KSHV miRNAs, leading the authors to hypothesize that the circRNAs derived from the human genome might sequester KSHV-derived miRNAs and prevent their repressive impact upon target mRNAs. The authors focused on hsa_circ_0001400, which is highly abundant in HUVECs and is moderately elevated following infection; by contrast, the levels of linear transcript RELL1 mRNA, which overlaps partly with hsa_circ_0001400, was not affected in KSHV-infected cells. While hsa_circ_0001400 levels did not change in the different viral phases, latent or lytic, its overexpression reduced the levels of viral LANA and RTA mRNAs, and its silencing elevated these mRNAs. In future work, it will be important to investigate other cellular circRNAs increasing after infection, as their function may synergize with the effects of hsa_circ_001400 in suppressing the expression of KSHV genes. Among a subset of interferon-stimulated proteins, only TNF was upregulated by hsa_circ_0001400, suggesting that the circRNA might function in antiviral host defense via TNF production. Given the clinical implications of the findings in this report, the possibility that TNF is an effector of hsa_circ_0001400 actions also warrants future investigation.
The identification of viral and mammalian circRNAs produced after KSHV infection raises many interesting questions, particularly regarding the mechanisms whereby KSHV circRNAs might promote virulence while mammalian circRNAs might prevent it (Figure 1). Analyzing the abundance, expression timeline, and localization of these circRNAs, as well as identifying en masse the interacting factors—proteins, RNAs, DNA, and other macromolecules—will shed critical light on the functions of circRNAs in mounting and counteracting KSHV infection. More broadly, the report by Tagawa et al. sets the stage for investigating the expression and function of circRNAs produced in response to other viral infections. Similar to other areas of circRNA biology, there is urgent need for global approaches to study circRNA sequences, copy numbers, associated factors, and possible translation profiles of circRNAs synthesized during the viral response.
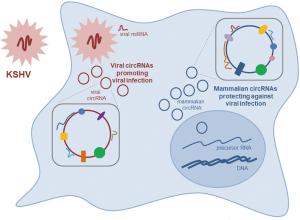
Finally, as suggested in the Tagawa report, some endogenous circRNAs may provide defense against viral infection. These circRNAs could be exploited therapeutically to prevent or reduce viral disease. For example, administration of hsa_circ_001400, possibly in combination with other cellular circRNAs induced by KSHV, might engender a more robust anti-viral response. Similarly, antagonization of KSHV-encoded circRNAs could have additional therapeutic benefit. Towards these goals, it is imperative that biological methods be developed to produce protective circRNAs and antagonizing RNAs at a large scale, as well as methods to direct them to target tissues for antiviral outcomes in the clinic.
Acknowledgments
The authors are funded by the National Institute on Aging Intramural Research Program, National Institutes of Health.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Chang Y, Cesarman E, Pessin MS, et al. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 1994;266:1865-9. [Crossref] [PubMed]
- Ablashi DV, Chatlynne LG, Whitman JE Jr, et al. Spectrum of Kaposi's sarcoma-associated herpesvirus, or human herpesvirus 8, diseases. Clin Microbiol Rev 2002;15:439-64. [Crossref] [PubMed]
- Cesarman E, Chang Y, Moore PS, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med 1995;332:1186-91. [Crossref] [PubMed]
- Soulier J, Grollet L, Oksenhendler E, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 1995;86:1276-80. [PubMed]
- Calabro ML, Sarid R. Human Herpesvirus 8 and Lymphoproliferative Disorders. Mediterr J Hematol Infect Dis 2018;10:e2018061. [Crossref] [PubMed]
- Pellett PE, Wright DJ, Engels EA, et al. Multicenter comparison of serologic assays and estimation of human herpesvirus 8 seroprevalence among US blood donors. Transfusion 2003;43:1260-8. [Crossref] [PubMed]
- Mosam A, Carrara H, Shaik F, et al. Increasing incidence of Kaposi's sarcoma in black South Africans in KwaZulu-Natal, South Africa (1983-2006). Int J STD AIDS 2009;20:553-6. [Crossref] [PubMed]
- Engels EA, Biggar RJ, Hall HI, et al. Cancer risk in people infected with human immunodeficiency virus in the United States. Int J Cancer 2008;123:187-94. [Crossref] [PubMed]
- Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology 2011;411:325-43. [Crossref] [PubMed]
- Campbell M, Kim KY, Chang PC, et al. A lytic viral long noncoding RNA modulates the function of a latent protein. J Virol 2014;88:1843-8. [Crossref] [PubMed]
- Toptan T, Abere B, Nalesnik MA, et al. Circular DNA tumor viruses make circular RNAs. Proc Natl Acad Sci U S A 2018;115:E8737-45. [Crossref] [PubMed]
- Panda AC, Grammatikakis I, Munk R, et al. Emerging roles and context of circular RNAs. Wiley Interdiscip Rev RNA 2017. [Crossref] [PubMed]
- Abdelmohsen K, Panda AC, De S, et al. Circular RNAs in monkey muscle: age-dependent changes. Aging (Albany NY) 2015;7:903-10. [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- Rybak-Wolf A, Stottmeister C, Glazar P, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 2015;58:870-85. [Crossref] [PubMed]
- Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014;15:409. [Crossref] [PubMed]
- Salzman J, Gawad C, Wang PL, et al. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7:e30733. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 2017;14:361-9. [Crossref] [PubMed]
- Tagawa T, Gao S, Koparde VN, et al. Discovery of Kaposi's sarcoma herpesvirus-encoded circular RNAs and a human antiviral circular RNA. Proc Natl Acad Sci U S A 2018;115:12805-10. [Crossref] [PubMed]
- Panda AC, De S, Grammatikakis I, et al. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res 2017;45:e116. [Crossref] [PubMed]


