
MicroRNA expression profile by next-generation sequencing in a novel rat model of contrast-induced acute kidney injury
Introduction
Intravascular iodinated contrast media (CM) are being widely injected in diagnostic and therapeutic procedures, which lead to an increased incidence of contrast-induced acute kidney injury (CI-AKI) (1). CI-AKI frequently develops in patients with diabetes and renal dysfunction, which is associated with higher mortality following coronary angiography (2).
The definite mechanism of CI-AKI is unknown. MicroRNA molecules (miRNAs) are a class of approximately 19-23 nucleotides non-coding RNAs. miRNAs have been reported to be associated with many kidney diseases including CI-AKI, but most of the miRNA research in kidneys has focused on identifying unique miRNA expression patterns in kidney diseases, such as a study which investigated miR-21 deteriorated diabetic kidney disease in mice (3). MicroRNA-30c-5p and microRNA-192-5p in urine may be potential biomarkers for ischemia-reperfusion-induced kidney injury (4). Our previous study showed that miR-21 antagonized cell apoptosis through PDCD4 in CI-AKI (5). Other recent research also examined the miRNA expression profile in plasma and kidney of CI-AKI rat model, but they injected a nonionic low-osmolar contrast medium, Omnipaque, which was not used in cardiac catheterization (6). Therefore, to gain a comprehensive understanding of miRNA expression in CI-AKI, we established a novel practical CI-AKI rat model by using a nonionic low-osmolar iodic contrast medium Ultravist is was frequently-used in cardiac catheterization and performed miRNA microarray assays, analyses of gene ontology (GO) categories, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
Methods
Novel CI-AKI rat model
Male Sprague-Dawley rats (200–220 g) were purchased from the Animal Center of Guangdong province (Guangzhou, China) and the model was accomplished in the Department of Experimental Animal Research Center, Southern Medical University (Guangzhou, China). The rats were acclimatized for 7 days before the start of the study, and the animal experiments in the present study were conducted accordance with the Guide for the Care and Use of Laboratory Animals as approved by the US National Institutes of Health (8th Edition, National Research Council, 2011) and our institutional and national guidelines for animal research; the protocol was approved by Guangdong General Hospital Ethics Research Committee.
Based on a previous method used for the CI-AKI rat model (7), we established a new method to induce our CI-AKI model. In brief, the rats were put under a 10% chloral hydrate (15 mL/kg) intraperitoneal anesthesia. Furosemide was administrated intraperitoneally at 15 mL/kg and dehydration for 6 hours before a nonionic low-osmolar contrast medium Ultravist (iopromide injection) (300 mg iodine/mL, 330 mosm/kg of water and 11.8 cPs at 37 °C, Bayer AG, Leverkusen, Germany) was injected at 15 mL/kg via tail vein administration over the course of 5 min. After contrast administration exposure, dehydration was sustained for 6 hours.
Sampling and assessment of rat renal functions
Blood samples were taken from the orbital vein at baseline and 24 h after contrast administration. Serum creatinine (Scr), blood urea nitrogen (BUN) and blood cystatin C (Cys C) concentrations were determined by a standard spectrophotometric assay using AU 5800 chemistry analyzer (Beckman Coulter, Inc. CA, USA).
Renal tissues collection
Twenty-four hours after contrast administration exposure, kidneys were excised and cut from the top to the bottom. Tissues were fixed in 10% neutral-buffered formalin and dehydrated in a graded series of alcohols. Then, the tissues were deparaffinized in xylenes and paraffin-embedded. After that, the 5 µm thick sections were cut from the paraffin blocks and routinely dewaxed and hydrated. The slices were stained with hematoxylin, rinsed with water, differentiated with 1% hydrochloric acid alcohol, stained with eosin for 1 minute and rinsed again with water. Finally, the slices were dehydrated with alcohol followed by red paraffinized in xylene and mounted with coverslips.
MicroRNAs microarray procedures
Sample preparation, library construction, and Illumina sequencing
Three panicles and 3 replicates of each tissues were used for RNA sequencing. Libraries were constructed using the Illumina Gene Expression Sample Preparation Kit and sequenced using the Illumina HiSeqTM 2000 (next generation sequencing) from BGI, Beijing Genomics Institute. Briefly, total RNA was isolated from the sample and first treated with DNase I to degrade any possible DNA contamination and then enriched for mRNA by using oligo (dT) magnetic beads. The enriched mRNA was mixed with fragmentation buffer and fragmented into short fragments (about 200 bp) from which the first strand was synthesized by using a random hexamer-primer. The second strand was synthesized by adding a reaction buffer, dNTP, RNaseH and DNA polymerase I to the first strand synthesis mixture. The double-stranded cDNA was purified with magnetic beads and then subjected to a 3'-terminal single nucleotide adenine. Finally, the sequencing adaptor is ligated to the fragment for PCR amplification. Enriched fragments were sequenced by Illumina HiSeqTM 2000 and 50 base pairs of raw reads were generated by Illumina Genome Analyzer II.
Data processing and bioinformatic analysis
The raw readings were filtered to obtain high quality reads by removing reads with an adaptor sequence or reads with a percentage of unknown bases (N) greater than 10%, and wherein the low-quality reads contained more than 50% bases and Q <5. The high-quality reads obtained were mapped to the rice reference genome (IRGSP build 5.0) using SOAPaligner/SOAP2; no more than two mismatches were allowed in the alignment. The False Discovery Rate (FDR) is used to determine the threshold of P values in multiple tests and analyses by manipulating FDR values (Benjamini & Yekutieli 2001). The absolute value of FDR ≤0.001 and log2-ratio ≥1 is used as a threshold for judging the significance of gene expression difference. We use the Blast2GO program to get the DEG’s GO annotation. Blast2GO is a widely recognized GO annotation software. The web-based software WEGO is used to classify GO functions for all annotated DEGs and to view macro-level gene function distributions using default parameters. The KEGG pathway is used to analyze the pathways involved in CI-AKI.
Quantitative real-time polymerase chain reaction (qRT-PCR)
qRT-PCR analysis was performed to confirm the accuracy of the microarray data. MiRNA expressions were determined on a vii A7 quantitative PCR system (Applied Biosystems, Carlsbad, CA, USA) and calculated using 2-ΔΔCt (Ct, threshold cycle) (n=8). The expression levels of several putative target genes were evaluated (n=8). U6 and β-actin were used as endogenous controls for miRNA and mRNA respectively. Primers for qRT-PCR were designed by the primer synthetic primer primer 5.0 and are listed in Table S2.
Total RNA was extracted from samples of rat kidney tissue and scrum using chloroform and TRIzol reagent (Invitrogen, Carlsbad, CA, USA). First strand cDNAs were generated from 1.5 µg of total RNA using a mixture of oligo(dT)15 and random primers or specific primers with superscript reverse transcriptase (Invitrogen, Carlsbad, CA, USA) according to the instructions. SYBR-based qRT-PCR (SYBR Premix Ex TaqTM II kit, TaKaRa code: DRR820A) was set in a reaction volume of 20 µL and repeated 3 times on a vii A7 quantitative PCR system (Applied Biosystems, Carlsbad, CA, USA). The following reaction conditions were used: 95 °C for 1 minute, then 40 cycles, 95 °C for 10 seconds and 60 °C for 30 seconds.
Analysis method
All statistical calculations were performed using SPSS 17.0 software (SPSS, Chicago, IL, USA). All data are expressed as mean ± standard deviation. The original signal intensity of the miRNA was calculated after a parallel reaction standardized to 5S RNA hybridization. Clustering and visualization of normalized data was performed using Cluster and TreeView (Michael Eisen, Stanford University, USA). The 2-ΔΔCt method is used to present relative gene expression. Comparison of miRNA expression between control and CI-AKI model mouse tissues was tested by Student’s t-test. A value of P<0.05 was considered to be significant.
Results
Contrast-induced renal dysfunction in rats
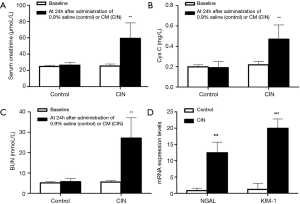
Rats were executed at 24 h after contrast administration exposure, and the renal function of rats was monitored too. Serum creatine and BUN of the CI-AKI group were increased about 2-fold change, more than 80% compared with baseline. Serum Cys C of the CI-AKI group was increased about 4-fold change. Kidney injury markers such as NGAL and KIM-1 also increase significantly. No significant alterations were observed in the control groups (P>0.05) (Figure 1).
Identification of differentially expressed miRNAs (DEMs)
The 50 nucleotide sequence tags from Hiseq sequencing were analyzed by data cleanup to obtain reliable, clean labels. Then, the length distribution of the cleaning label and the common and specific sequence between the samples are summarized. Next, standard analysis annotates clean labels to different categories and those who cannot be annotated to any category predict new miRNAs and seeds to edit potentially known miRNAs. The whole process is shown in Figure 2A.
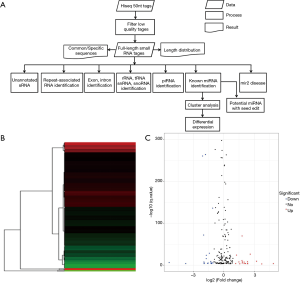
DEMs were identified by default settings via the DESeq package. DEM is defined when log2-Ratio (CI-AKI/control) ≥0; meanwhile, the FDR is set to 0.01. The DEGs of the library constructed from the CI-AKI group and the control group were compared, and a total of 207 unique genes were differentially regulated in the CI-AKI group. Of these, 100 were up-regulated and 107 were down-regulated (Figure 2B,C), from which 41 were significantly down-regulated (log2-ratio <−1, P<0.01) and up-regulated miRNA (log2-ratio >1, P<0.01) for further study (Table 1).
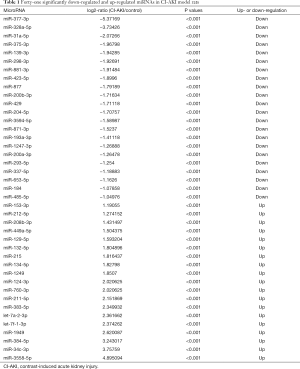
Full table
Functional enrichment analysis of putative target genes
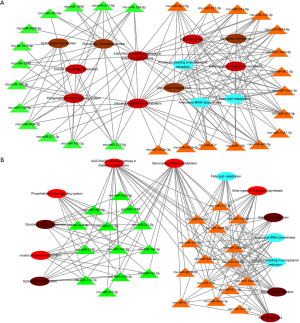
Target prediction was performed in 41 significantly down-regulated and up-regulated miRNAs. miRanda, RNAhybrid (Figure 3), and putative target genes were obtained from the intersection of the results between miRanda and RNAhybrid, and then analyzed by KEGG pathway and GO category. Putative genes were enriched in biological processes or signaling pathways related to AKI pathogenesis, including inflammation, apoptotic process and endoplasmic reticulum stress, along with genes implicated in signaling pathways involved in inflammation and dysfunctional metabolism, including AGE-RAGE signaling pathway and glycerophospholipid metabolism (FDR corrected, P<0.05) (Figure 3).
Validation of RNA-Seq-based gene expression by qRT-PCR
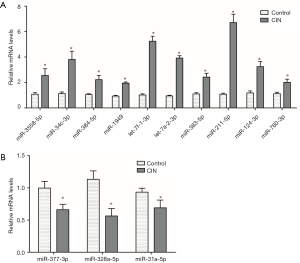
Thirteen DEMs including 3 down-regulated DEMs (miR-377-3p, miR-328a-5p, miR-31a-5p) and 10 up-regulated DEMs (miR-3558-5p, miR-34c-3p, miR-384-5p, miR-1949, let-7a-2-3p, let-7f-1-3p, miR-383-5p, miR-211-5p, miR-124-3p, miR-760-3p) out of the 41 significantly down- or up-regulated DEMs were selected (log2-ratio ≥2, P<0.01) for qRT-PCR. RNA samples are collected independently in order to verify the gene expression level predicted by RNA-Seq. Results (Figure 4) demonstrated that the detected results by qRT-PCR are consistent with the predicted results.
Discussion
Our next generation sequencing showed that of the 173 miRNAs 22 were significantly down-regulated (miR-377-3p, miR-328a-5p, miR-31a-5p, et al.) and 19 were significantly up-regulated (miR-3558-5p, miR-34c-3p, miR-384-5p, et al.) in the kidneys of CI-AKI rat model, which included new DEMs, such as miRNA-1949 and miR-3558. The results showed that a large set of genes involved in essential biological processes were targeted by these miRNAs, such as metabolic dysfunction process, apoptotic process and endoplasmic reticulum stress, along with genes implicated in signaling pathways involved in inflammation and dysfunctional metabolisms, such as AGE-RAGE signaling pathway and glycerophospholipid metabolism.
The occurrence of CI-AKI after percutaneous endovascular procedures has been associated with several factors, such as patient baseline characteristics, procedural variables and confers unfavorable prognosis. It has been demonstrated that the risk of CI-AKI and its harmful consequences are present in patients with or without chronic kidney disease and increase in diabetic patients (8). However, other reported CI-AKI risk factors have not been validated and practical convenient, fast methods to assess CI-AKI risk in patients undergoing percutaneous endovascular procedures have not been developed yet.
Ben at also investigated the miRNA profile in renal tissue in CI-AKI rat models and found that plasma levels of candidate miRNAs such as miRNA-188, miRNA-30a, and miRNA-30e can serve as potential biomarkers for early (4 hours) detection of CI-AKI; this finding was not validated in our novel CI-AKI rat model. Our study adopted the frequently-used contrast, a nonionic low-osmolar contrast medium Ultravist (iopromide injection), while Ben’s study adopted another nonionic low-osmolar contrast medium Omnipaque (9). In addition, they injected contrast medium and furosemide in a lower dose (10 vs. 15 mL/kg) and used different technology (miRNA arrays vs. next generation sequencing); despite this, we also found similar results including down-regulated DEMs (miR-139-3p, miR-429, miR-204-5p, miR-200b-3p) and up-regulated DEMs (miR-212-5p and miR-760-3p). Unfortunately, our present study has not validated plasma miRNA profile in rats and human, nor has it done so in ex vivo or animal model.
Gutiérrez-Escolano et al. established a rat model of CI-AKI involving dehydration, followed by administration of furosemide (10 mg/kg), iohexol (10 mg/kg), and indomethacin (10 mL/kg), which may have increased renal toxicity and confounders (10). Their microarray assay (kidneys tissue) showed 7 of 51 miRNAs having a >2.0-fold change in expression, including elevated expression levels in miR-21, miR30 family, et al. and down-regulated miRNAs such as miR-200a, and miR-204, which were validated in our novel model.
Our previous study showed contrast exposure induced cell apoptosis and decreased miR-21 expression in the kidney tissues of CI-AKI rat model. After transfection of miR-21 mimics, the expression of programmed cell death protein 4 (PDCD4) was inhibited and apoptosis was antagonized. However, our study did not find the differentially expressed miR-21 in the novel animal model of CI-AKI.
In this study, new DEMs, such as miRNA-1949, miR-3558, miR-1247 and miR-3594 were identified in the injured kidney tissues. Through analyses of GO categories and KEGG pathways, genes were implicated in signaling pathways involved in inflammation and dysfunctional metabolisms, such as AGE-RAGE signaling pathway and glycerophospholipid metabolism. Whether the DEMs/AGE-RAG pathway exhibits this function in vivo will be investigated in future studies.
Acknowledgements
Funding: This study was supported by the National Science Foundation for Young Scientists of China (grant no. 81500520); the National Science Foundation of China (grant no. 81670339); Science and Technology Planning Project of Guangdong Province (grant no. 2015A030302037); Science and Technology Planning Project of Guangdong Province (grant no. 2014B070706010); Guangdong Provincial Medical Research Fund Project (grant no. GSIC20140526); and Guangdong Provincial People’s Hospital Clinical Transformation Research Project (grant no. 2015zh01).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The protocol was approved by Guangdong General Hospital Ethics Research Committee (No. 2015253).
References
- Nash K, Hafeez A, Hou S. Hospital-acquired renal insufficiency. Am J Kidney Dis 2002;39:930-6. [Crossref] [PubMed]
- Pannu N, Wiebe N, Tonelli M, et al. Prophylaxis strategies for contrast-induced nephropathy. JAMA 2006;295:2765-79. [Crossref] [PubMed]
- Li R, Chung AC, Yu X, et al. MicroRNAs in Diabetic Kidney Disease. Int J Endocrinol 2014;2014:593956. [Crossref] [PubMed]
- Karbiener M, Neuhold C, Opriessnig P, et al. MicroRNA-30c promotes human adipocyte differentiation and co-represses PAI-1 and ALK2. RNA Biol 2011;8:850-60. [Crossref] [PubMed]
- Wang K, Bei WJ, Liu YH, et al. miR-21 attenuates contrast-induced renal cell apoptosis by targeting PDCD4. Mol Med Rep 2017;16:6757-63. [Crossref] [PubMed]
- Sun SQ, Zhang T, Ding D, et al. Circulating MicroRNA-188, -30a, and -30e as Early Biomarkers for Contrast-Induced Acute Kidney Injury. J Am Heart Assoc 2016;5. [Crossref] [PubMed]
- Ulusoy S, Ozkan G, Mungan S, et al. GSPE is superior to NAC in the prevention of contrast-induced nephropathy: might this superiority be related to caspase 1 and calpain 1? Life Sci 2014;103:101-10. [Crossref] [PubMed]
- Zuk A, Bonventre JV. Acute Kidney Injury. Annu Rev Med 2016;67:293-307. [Crossref] [PubMed]
- Sun S, Zhang T, Nie P, et al. A novel rat model of contrast-induced acute kidney injury. Int J Cardiol 2014;172:e48-50. [Crossref] [PubMed]
- Gutiérrez-Escolano A, Santacruz-Vázquez E, Gómez-Pérez F. Dysregulated microRNAs involved in contrast-induced acute kidney injury in rat and human. Ren Fail 2015;37:1498-506. [Crossref] [PubMed]


