
Genetics and epigenetic factors of Wilson disease
ATP7B gene, protein structure and function
Wilson disease (WD) is considered a monogenic autosomal recessive disease and ATP7B is the only gene that has been identified as causative of this condition. Disease-causing mutations affecting the ATP7B gene are associated with lack of biliary copper excretion and its consequent accumulation in the hepatocytes. The ATP7B gene encodes for a transmembrane P-type cation copper transporter ATPase and can be affected by homozygous or compound heterozygous mutations, meaning that each gene allele presents a different mutation. The gene is located on human chromosome 13q14, it contains 20 introns and 21 exons, and almost 800 mutations have been described according to the Human Gene Mutation Database (accessed January 2019) (1-3). Not all mutations are causative of copper accumulation. The clinical phenotype and the relation between ATP7B gene mutations with the extent of the copper transporter function impairment are not predictable (4). Therefore, even the most advanced technologies for genetic diagnosis, including next generation sequencing, still require evidence of abnormal copper metabolism parameters for the definite clinical diagnosis of WD (5). There have been reports of pseudo-dominant inheritance where the disease presents in 2 or more consecutive generations within one family (6,7). Rarely, one individual can present with three mutations. Cases of segmental uniparental disomy, occurring when both copies of a single chromosome are inherited from one parent, have been described (8).
The ATP7B gene is highly expressed in the liver but the protein is also detected in the brain, kidney, lung, intestinal epithelial cells, placenta, and mammary glands (9). The gene encodes for a 1465-aminoacid protein which is synthetized in the endoplasmic reticulum and is expressed in the trans-Golgi network (10). The ATP7B protein belongs to class 1B (PIB) P-type ATPase superfamily, responsible for actively exporting copper and other heavy metal ions from the cytoplasm of cells. These highly conserved structure proteins present a phosphatase domain (A-domain), a phosphorylation domain (P-domain), a nucleotide-binding domain (N-domain), and M-domains, which includes eight transmembrane ion channels. The copper transport through the membrane is an active complex mechanism which is ATP-dependent and functionally affected by the gene mutations. Many of the described mutations map at the N-domain, potentially impairing the transporter function by interfering with the ATP binding (11,12).
ATP7B gene mutations and population prevalence
More than 50% of the described mutations are single-nucleotide missense and nonsense, whereas the remaining are insertions/deletions and splice site mutations. The missense mutation His1069Gln on exon 14 is the most prevalent in patients of Central and Eastern European descent (13). However, its prevalence varies greatly according to the geographic area. The allele frequency is the highest in Poland, Latvia, Former East Germany, Bulgaria, and Czeck Republic ranging from 72% to 57% (14-18). The mutation is also the one with highest prevalence in North America with about 40% allele frequency (19). A study on a large group of Polish patients with WD using whole-exome sequencing, confirmed the high prevalence of the H1069Q mutation and identified other low-prevalence mutations (20). Another described mutation in Central and Eastern Europe is the G710S on exon 8, having a prevalence of 6.4% in Austria (21). In United Kingdom, the most frequent mutation is the H1069Q but with a prevalence of 19%, much lower compared to other European countries (8). The second most common identified mutation in UK is the Met769Val with a frequency of 6% (8). Patients from continental Italy present a lower prevalence of the H1069Q mutation (17.5%) and other mutations (p.Val845fs and p.Met769fs) are described with a prevalence lower than 10% (22). Spain represents an exception compared to other European countries with a high frequency of the missense mutation M645R on exon 6 (23). A study on French patients with WD also identified the M645R as highly prevalent with a carrier frequency of 1.8% (5). Regions where consanguinity is highly prevalent, present different patterns of mutations distribution. Sardinia has a high prevalence (60%) of the deletion c.-441_-427del15 in the ATP7B gene promoter region (24) and Canary Island has a 64% allele frequency of the p.Leu708Pro (25). In Costa Rica, patients present with a high prevalence (61%) of the N1270S mutation (19), also described in Italian and Turkish populations (26,27). In China, Japan, and Korea the prevalent mutation is the missense R778L on exon 8 with an allele frequency ranging from 45% to 12% (28-31). In Japan, the 2871delC and 2874delC are also described (32). This variant is associated with replacement of arginine with leucine in the transmembrane domain. India is a vast country with high ethnic diversity and different prevalence of ATP7B mutations has been described according to the geographic localization. As pointed out by Gomes et al, the p.Cys271 mutation appears to be the most frequent mutation in the Indian sub-continent (33,34) even though it has not been described in the north of the country.
ATP7B mutations, copper transporter function, and genotype-phenotype correlation
Whereas the pathogenesis of WD is known to be rooted in copper accumulation and ATP7B copper transporter impaired function, it is much less clear if and how ATP7B mutations influence the phenotype. At the molecular level, previous studies have shown a relationship between the type of gene mutation and the extent of copper transporter functional impairment. A study has characterized in vitro and in silico the activity changes induced by the H1069Q mutation showing that its main consequences are protein misfolding, aberrant phosphorylation of the P-domain, and altered ATP binding orientation and affinity (35). Another study using immunogold electron microscopy to explore the intracellular localization of ATP7B in liver tissue from WD patients homozygous for the H1069Q mutation, identified an aberrant localization of the mutated protein in the endoplasmic reticulum whereas the non-mutated ATP7B was in the trans-Golgi network (36). An extensive in vitro study assessed the catalytic and transport activity as well as the intracellular localization of 28 ATP7B variants. The selected variants, including the H1069Q, were all known to be associated with clinical manifestations. Various mutations were associated with different functional impairments but notably variants domain-localization was not the cause of the same type or extent of dysfunction (37). Common missense mutations tend to impair ATP7B stability and are associated with its reduced levels (38,39). In addition, several mutations impair only partially ATP7B function with some residual ATP binding and copper transporter activity (36). There is evidence that certain ATP7B polymorphisms may interact with metabolic factors, specifically copper availability, with effects on the copper transporter activity itself. In particular, the Arg875 causes lack of copper transporter activity but in response to elevated copper, the trafficking activity was restored along with the copper delivery (40). A study on 59 patients with WD found that protein-truncating mutations were associated with lower serum ceruloplasmin oxidase activity compared to missense mutations (41). A study from India identified 28 ATP7B variants in 50 patients with WD and conducted in vitro functional analysis describing 5 new variants associated with protein misfolding (34). When transitioning from the molecular to the disease level, several studies embarked in the difficult task to identify a correlation between genotype and phenotype but the results have been non-conclusive and affected by several challenges. First, most patients are compound heterozygous. Second, the clinical practice (for example, hepatology versus neurology) could have influenced the initial selection of cases and in some cases, liver or neurological signs or symptoms may have not been properly investigated or diagnosed. Third, most studies include a overall small number of cases, assessed and diagnosed with non-standardized criteria, often uniquely based on the single clinician experience. The most recent and relevant evidence of a limited role of the genotype on the phenotypic manifestations is provided by a European study on more than 1300 patients with WD, including 702 children and 655 adults. Patients presented either hepatic or neurological manifestations and more than 50% of them had a liver biopsy-confirmed diagnosis. The described population included both acute liver failure and cirrhosis patients carrying a high allele frequency of the H1069Q mutation (46.9%) (42). The major finding of the study was the lack of correlation between ATP7B gene mutations and the phenotype. It should be mentioned that this study was preceded by multiple other smaller studies attempting to identify a genotype-phenotype correlation. In particular, the H1069Q mutation was associated more frequently with neurological phenotype in some studies (16,43). A Dutch study showed an association between homozygous H1069Q and neurological phenotype (44). A study on 58 pediatric patients with WD affected by 34 different ATP7B mutations showed that nonsense and frameshift mutations were associated with lower serum ceruloplasmin and copper levels (45). Genotype-phenotype correlation studies in 126 Bulgarian patients presenting a H1069Q allele frequency in 78% of cases indicated an association between the mutation and hepatic presentation (46). Tarnacka et al. reported on 148 Polish patients with a high H1069Q frequency and did not find any genotype-phenotype correlation (47).
Other interesting information can be derived from studies describing siblings and homozygous twins. Chabik et al. studied 73 unrelated Polish families including 73 index cases and 95 siblings, presenting with a H1069Q allelic frequency of 77% (48). They found a high concordance rate of the initial clinical presentations, being 86% concordance for hepatic symptoms and 66% for neurological symptoms. However, there are also case reports of siblings and homozygous twins presenting discordant clinical presentations or different post liver transplant outcomes (49-51). A Japanese study in 11 families including 23 sibilings diagnosed with WD described that 5 families had identical phenotypes and 6 families different phenotypes (52). Interestingly, there is evidence of geographical clustering within regions with smaller or isolated communities with various levels of consanguinity and presenting phenotype homogeneity. Two large families in small mountain community in the region of Rucar in Romania were studied given the high prevalence of WD cases (53). Of the 50 screened living members, 5 individuals with clinical diagnosis of WD and 2 asymptomatic subjects presented the H1069Q/M769H mutations. Of note, there was a significant phenotypic concordance between all WD patients presenting neurological and psychiatric phenotype. Patients presented mostly dysarthria and dysphagia as initial symptoms and had similar age of onset (17–20 years old). The authors concluded that their findings demonstrate the influence of both genetic and environmental factors on the phenotype (53). More than 70 members of a single Lebanese family were investigated for the c.2299insC and p.Ala1003Thr mutations. The clinical diagnosis of WD was confirmed in 9 subjects (54). The c-2299insC mutation was associated hepatic and the Ala1003Thr was associated with neurological phenotype. A study on 4 generations of an Italian family, showed a possible association between the homozygotic mutation T1288R and the hepatic phenotype (55). Different evidence comes from Gran Canaria, where the Leu708Pro mutation was highly prevalent, affecting 18 out of 24 patients with WD, including 12 homozygous patients, 4 compound heterozygous, and 2 with only one identified mutation (25). However, the phenotype was variable and included prevalently neurological manifestations but also hepatic involvement with no obvious association between the genotype and phenotype. Therefore, despite the methodological challenges, the evidence points to the presence of additional factors, other than gene mutations, affecting the clinical presentation.
Modifier genes
A modifier gene has been defined as a gene that alters the expression of a gene at another locus or the phenotypic expression of another gene (56). It is plausible to hypothesize the presence of modifier genes influencing the varied WD phenotype. Several genes have been proposed as possible candidates for this role. Kluska et al. conducted whole-exome sequencing analysis on 248 patients with WD (20). The study identified two new possible variants in esterase D (ESD) and INO80 genes being associated with increased and reduced risk of neurological presentation, respectively. ESD, encoding for a polymorphic red cell enzyme, was previously linked to WD (57). INO80 has important transcription regulation functions through chromatin remodeling action (58). In addition, rare APOE and MBD6 variants were associated with lower risk of early onset WD (20). Other studies have previously associated APOE variants with WD (59), in one case with delayed neurological symptoms (60). However, other studies could not confirm the association (61,62). Interestingly, MBD6 encodes for a protein belonging to the methyl-CpG-binding domain family. These proteins have central regulating function in epigenetic mechanisms, including the readout of DNA methylation (63). MBD6 interacts with the human deubiquitinase complex and reported to be a target of Oct4 in stem cells derived from adipose tissue (64). Conversely, the analysis could not confirm any association between WD phenotypes and other previously proposed modifier genes allelic variants (20). Even though the study results may have been affected by the relatively small population or by the analysis methodologies, the results question the validity and clinical significance of previous findings on candidate modifier genes.
A candidate modifier gene with mechanistic relevance is the patatin-like phospholipase domain-containing 3 gene (PNPLA3). PNPLA3 rs738409 polymorphism has been associated with increased risk of nonalcoholic fatty liver disease development (65) and with hepatic steatosis in hepatitis B (66) and C (67). The protein presents various functions in lipids metabolism as it can both synthetize intracellular triglycerides and also has hydrolyzing activity against triglycerides (68,69). Therefore, mutations affecting its function could favor steatosis. A study on 98 male patients with WD showed that, on multivariate logistic regression, PNPLA3 was an independent variable associated with moderate/severe steatosis (70). MTHFR mutations are also potential genetic modifiers of WD. MTHFR encodes 5,10-methylenetetrahydrofolate reductase, an enzyme in methionine metabolism that affect homocysteine levels. Two polymorphisms in MTHFR were associated with WD expressivity. The MTHFR 667T allele was associated with hepatic phenotype and the MTHFR 1298C allele was associated with earlier WD presentation (71). This is interesting as it provides evidence that aberrant homocysteine and methionine metabolism may interact with copper accumulation in the pathogenesis of WD and provides a possible explanation for the variable expressivity of this monogenetic disease.
Other previously proposed modifier genes for WD include antioxidant 1 copper chaperone (ATOX) (72), copper metabolism domain-containing 1 (COMMD) (73-75), X-linked inhibitor of apoptosis (XIAP) (76), hemochromatosis gene (HFE) (77-79), human prion gene (PrP) (80), ATP7A (81), and divalent metal transporter1 (DMT1) (81) but none was confirmed in additional or larger studies (Table 1).
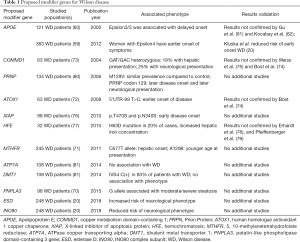
Full table
Introduction to epigenetics
The field of epigenetics, literally meaning “on top of genetics”, lies at the critical intersection where nature meets nurture, allowing a static genome to adapt to a dynamic environment. Epigenetics can explain long-lived effects from in utero and early life environments on human disease susceptibility and resilience. The “Developmental Origins of Adult Disease” hypothesis (82) relies heavily of epigenetic mechanisms to explain how maternal in utero effects can influence risk for common diseases in adulthood, including obesity, cancer, cardiovascular disease, addiction, and other mental health disorders (83,84).
Epigenetic information is layered on top of the DNA sequence through covalent modifications to nucleotides or nucleosomes (85,86). The first layer of epigenetic information is DNA methylation. The majority of CpG (meaning cytosine preceding a guanine) dinucleotides in the human genome are methylated at the 5’ position of cytosine. In contrast, CpGs are predominantly unmethylated when they are located in “CpG islands” at the promoters of approximately 70% of mammalian gene promoters. DNA methylation patterns are dynamically erased and reestablished each generation during the formation of the gametes, which occurs in utero in mammals (87). Implantation of the embryo is another critical time for genome-wide changes in DNA methylation patterns that can be modified by maternal environment. Adding to the complexity of DNA methylation dynamics, there are variations on 5-methylcytidine (mC), including 5-hydroxymethylation (hmC), which is correlated with gene activity and demethylation (88). Furthermore, while CpG dinucleotides are the most methylated, the more abundant CpH (C preceding an A, T, or C) dinucleotides are methylated at low frequency in mature oocytes, embryonic stem cells, and postmitotic neurons.
While DNA methylation has historically been considered to be a mechanism for silencing gene expression, the genome-wide view has demonstrated multiple complexities to that simple interpretation. 5-methycytosine at CpG sites at gene promoters is associated with gene repression. In contrast, a positive association with expression and CpG methylation is observed over gene bodies, particularly in pre-implantation embryonic tissues and placenta (87,89,90). Interpretation of CpH methylation is less clear, although it appears to mark genes that were active in earlier development that were later silenced (91,92). In these ways, the layer of DNA methylation can provide important clues about past, present, and future gene expression patterns.
In addition to DNA methylation, there are several other layers of epigenetic information. DNA in the nucleus is organized and assembled through the action of nucleosomes, which are octamers of pairs of four histone core subunits (H2A, H2B, H3, H4) and the H1 linker histone. All of the core histones have N- and C-terminal tails that are post-translationally modified by methylation, phosphorylation, ubiquitinylation, acetylation, to name a few (93). A large number of nuclear proteins in mammalian genomes are dedicated to the writing, reading, or erasing of these histone modifications collectively known as the “histone code”. Genome-wide, histone codes have been mapped for a large variety of human tissues, developmental stages, and cell types in order to functionally annotate the genome into promoters versus enhancers, active versus repressed versus “poised” genes, as well as inactive heterochromatin. Chromatin loops between enhancers-promoter pairs, or groups of genes with a single “super-enhancer” have also been mapped in specific cell types and tissues genome-wide (94). Lastly, noncoding RNAs are emerging as having a variety of functional roles in defining chromatin states and chromatin loop interactions between promoters and enhancers (95).
Integration of methionine metabolism with epigenetics and copper
There is a large variability in the expressivity of WD in terms of the age of onset, severity, response to treatment, and impacts to the liver versus brain. Analysis of hundreds different disease-causing mutations in WD have failed to find convincing evidence of genotype-phenotype correlations. Therefore, WD appears to be a genetic disease with clear environmental and epigenetic modifications that are likely due to the complex relationship between copper accumulation and methionine metabolism pathways (96). Methionine metabolism in liver and brain is an important pathway that regulates the supply of methyl groups required for modifications of proteins, DNA, and RNA through the availability of the universal methyl donor S-adenosylmethionine (SAM). Methionine adenosyltransferase utilizes ATP to convert methionine into SAM. S-adenosylhomocysteine hydrolase (AHCY) is a key enzyme regulating the amount of SAM available for methylation reactions, since it bidirectionally catalyzes the conversion of SAM to S-adenosylhomocysteine (SAH). Bethin et al. initially demonstrated that in toxic milk mouse livers AHCY has copper binding properties and the enzymes levels were reduced by 42% compared to mice with normal copper metabolism (97). Subsequent studies demonstrated that excess copper inhibits the activity of AHCY (98). AHCY inhibition leads to reduced SAM/SAH ratios, inhibition of MAT, and impaired methylation reactions. Therefore, some of the variability of WD symptoms is likely to be due to wide-scale alterations in methyl donor supplies.
Studies on the toxic milk (tx-j) mouse model of WD are consistent with methionine metabolism alterations due to copper accumulation. Specifically, tx-j mice show reduced AHCY transcript and protein levels and elevated SAH and SAH/SAM ratios compared to wild-type mice (99). The methyl donor nutrient betaine counteracted the genetic effect, demonstrating further evidence that the tx-j phenotype could be modified by dietary methyl donors (99). Similar data in Long-Evans Cinnamon rats demonstrated a correlation between hepatic copper accumulation and reduced transcript levels of MAT and changes in methionine metabolism (100).
Integration of copper with mitochondrial oxidative stress and epigenetics
Another way that excess copper levels in WD may impact epigenetic layers is through the mitochondrial oxidative stress pathways. The intracellular levels of reactive oxygen species (ROS) increase as a byproduct from mitochondrial activity during normal cellular metabolism. While ROS are signal transduction molecules that are part of normal metabolism, high levels of ROS can cause detrimental effects on multiple cell components, including DNA, lipids, proteins, membranes, and mitochondria. ROS and additional mitochondrial metabolites have a detrimental impact on epigenetic layers, including DNA methylation and histone modifications such as methylation and acetylation (101,102).
Exposure to non-physiological levels of trace elements such as copper contributes to mitochondrial dysfunction, including oxidative stress. Other trace metals with an observed epigenetic impact in mammalian tissues, include nickel and iron. Interestingly, iron acts as a cofactor for the TET family proteins that catalyze the oxidation of DNA methylation from methylated to hydroxymethylated cytosine, while nickel acts as an inhibitor of the same reaction (103). Furthermore, nickel, chromium, and arsenic appear to be mechanistically involved in alterations to histone post-translational modifications by altering actions of the enzymes or “writers” that modify them (104). Therefore, the impact on the epigenome from even mild alterations to metals may have both direct effects on epigenetic enzymes or indirect effects through the mitochondrial and oxidative stress pathways.
Evidence for epigenetic and methionine alterations in animal models of WD
Since alterations in methionine metabolism can directly affect methylation reactions to DNA and histone tails, the copper-induced alterations to methionine pathways can also adversely impact many downstream gene targets through epigenetics. The tx-j mouse model of WD supports epigenetic alterations in WD. An initial time-course study on tx-j mice livers from fetal life to adulthood, showed global DNA methylation starting at 20 weeks of age with a significant negative correlation with Dnmt3a and Dnmt3b transcript levels (105). Pooling data from 20 and 28 week old mice, global DNA methylation correlated negatively with hepatic copper concentration and with SAH levels and correlate positively with SAM/SAH ratio. Tx-j liver samples show reduced Dnmt3b transcript levels and global DNA methylation levels compared to wild-type that were able to be restored through betaine supplementation (105). In addition to methionine, mitochondrial pathways directly impacted by excess copper accumulation result in ROS and other metabolites that influence epigenetic control of gene expression throughout the genome (106).
Epigenetic mechanisms are especially important in utero, because this is a critical time when the instructions of the genome are being followed within the context of the maternal environment, diet, and metabolism. In WD, this is particularly evident in the tx-j mouse model, as choline supplementation to the diet of dams from 2 weeks during pregnancy to gestational day 17 resulted in a 17% increase in global DNA methylation levels in the liver (107). Maternal choline supplementation also prevented the transcriptional deficits in fetal tx-j liver for multiple genes related to cell growth and rescued reduced body weight phenotypes of tx-j mice (107). Interestingly, copper levels in fetal livers of tx-j mice are actually lower than wild-type, which is the opposite pattern of the copper accumulation in postnatal livers of tx-j mice. What the two stages have in common is deficits in copper transport due to Atp7b mutation, as well as alterations to DNA methylation observed in liver.
Recently, we have applied genome-wide approaches to identifying the genes impacted by epigenetic changes in fetal livers of tx-j mice, as well as the effect of maternal choline supplementation (108). Using a whole methylome approach called whole genome bisulfite sequencing, we identified six differentially methylated genes with genome-wide significance, including Atp7b itself, as well as genes involved in oxidative stress and thioredoxin, including Gpx4, Prdx2, and Hif1a. Importantly, we also demonstrated that maternal choline supplementation corrected the tx-j differential methylation patterns observed. Secondly, by performing genome-wide analyses of transcriptional differences in the tx-j liver, we were able to demonstrate several associations between methylation and transcriptional changes in tx-j liver with and without choline supplementation (108). Together these results provided genome-wide evidence for epigenetic alterations in the tx-j model of WD that could be corrected through maternal choline supplementation.
Evidence for epigenetic alterations in samples from patients with WD
Unlike mouse model studies of WD in which the genetics and dietary methyl donors can be controlled, human studies have been more challenging to assess the role of epigenetics. However, genome-wide methylome sequencing approaches have been recently utilized in order to gain important insights into epigenetic changes in human WD liver and blood. We utilized whole-genome bisulfite sequencing (WGBS) to identify differentially methylated regions that distinguished liver biopsies from WD patients compared to both healthy and disease control subjects with other liver diseases. 18 regions at genome-wide significance were identified, as well as thousands of others at lower confidence, that were able to discriminate a WD-specific epigenomic signature. WD-specific differentially methylated regions were enriched for liver-specific enhancers and genes with functions in folate and lipid metabolism and acute inflammatory response.
We then sought to determine if blood samples from WD patients with hepatic versus neurologic symptoms could be distinguished by WGBS epigenomic signatures. While the differentially methylated regions identified were fewer than those identified in blood, there was a significant overlap (31% also in liver) with the >200 regions that distinguished WD blood DNA from healthy control or disease control samples. More importantly, differentially methylated regions in WD blood were identified that clearly discriminated those patients with hepatic versus neurologic symptoms in two different cohorts. These preliminary results suggest that epigenetic biomarkers in blood could be useful clinically in predicting the symptom and treatment types for early stage WD patients. To explore this possibility, we developed a classifier algorithm that predicted neurologic versus hepatic phenotypes in a separate cohort with 90% accuracy. Together these results suggest the potential clinical utility of mapping epigenetic alterations in WD human patients (109).
Sex and epigenetic differences
WD clinical presentations can be different between males and females (110,111) and the potential explanation could be related to epigenetics mechanisms Acute liver failure presents as WD onset more frequently in females whereas the neurological phenotype tends to manifest more in male patients. The explanation for these differences has not been completely clarified but in general has been attributed to hormonal influences or differences in iron metabolism (112). Hormonal factors are most likely involved but it is also likely that epigenetics factors are contributing to sex differences. Methylome and transcriptome differences have been described in human liver and brains and are associated with metabolic differences (113). Our studies in tx-j mice showed that female mice presented higher hepatic copper concentration compared to male mice after treatment with copper chelator penicillamine and global hepatic DNA methylation was different between male and female mice after treatment with choline (106). Moreover, female mice showed changes of hepatic transcript levels of genes related to oxidative phosphorylation in response to choline and penicillamine treatments (106).
Conclusions and questions for future research
Disease-causing mutations affecting the ATP7B gene cause a wide variety of functional alterations of the copper transporter. Genetic mutations are ultimately responsible for copper accumulation but other factors affect the clinical presentation. The interactions between genetic mutations and epigenetic factors may be the explanation for the heterogeneous phenotypic presentation of WD.
Acknowledgements
Funding: This work was supported by the National Institute of Health (DK104770 to V Medici).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Bull PC, Thomas GR, Rommens JM, et al. The Wilson disease gene is a putative copper transporting P-type ATPase similar to the Menkes gene. Nat Genet 1993;5:327-37. [Crossref] [PubMed]
- Petrukhin K, Fischer SG, Pirastu M, et al. Mapping, cloning and genetic characterization of the region containing the Wilson disease gene. Nat Genet 1993;5:338-43. [Crossref] [PubMed]
- Tanzi RE, Petrukhin K, Chernov I, et al. The Wilson disease gene is a copper transporting ATPase with homology to the Menkes disease gene. Nat Genet 1993;5:344-50. [Crossref] [PubMed]
- Czlonkowska A, Litwin T, Dusek P, et al. Wilson disease. Nat Rev Dis Primers 2018;4:21. [Crossref] [PubMed]
- Collet C, Laplanche JL, Page J, et al. High genetic carrier frequency of Wilson's disease in France: discrepancies with clinical prevalence. BMC Med Genet 2018;19:143. [Crossref] [PubMed]
- Dziezyc K, Gromadzka G, Czlonkowska A. Wilson’s disease in consecutive generations of one family. Parkinsonism Relat Disord 2011;17:577-8. [Crossref] [PubMed]
- Bennett JT, Schwarz KB, Swanson PD, et al. An exceptional family with three consecutive generations affected by Wilson disease. JIMD Rep 2013;10:1-4. [PubMed]
- Coffey AJ, Durkie M, Hague S, et al. A genetic study of Wilson's disease in the United Kingdom. Brain 2013;136:1476-87. [Crossref] [PubMed]
- Chang IJ, Hahn SH. The genetics of Wilson disease. Handb Clin Neurol 2017;142:19-34. [Crossref] [PubMed]
- Gourdon P, Liu XY, Skjorringe T, et al. Crystal structure of a copper-transporting PIB-type ATPase. Nature 2011;475:59-64. [Crossref] [PubMed]
- Barry AN, Shinde U, Lutsenko S. Structural organization of human Cu-transporting ATPases: learning from building blocks. J Biol Inorg Chem 2010;15:47-59. [Crossref] [PubMed]
- Inesi G, Pilankatta R, Tadini-Buoninsegni F. Biochemical characterization of P-type copper ATPases. Biochem J 2014;463:167-76. [Crossref] [PubMed]
- Gomes A, Dedoussis GV. Geographic distribution of ATP7B mutations in Wilson disease. Ann Hum Biol 2016;43:1-8. [Crossref] [PubMed]
- Gromadzka G, Schmidt HH, Genschel J, et al. Frameshift and nonsense mutations in the gene for ATPase7B are associated with severe impairment of copper metabolism and with an early clinical manifestation of Wilson’s disease. Clin Genet 2005;68:524-32. [Crossref] [PubMed]
- Krumina A, Keiss J, Sondore V, et al. From clinical and biochemical to molecular genetic diagnosis of Wilson disease in Latvia. Genetika 2008;44:1379-84. [PubMed]
- Caca K, Ferenci P, Kuhn HJ, et al. High prevalence of the H1069Q mutation in East German patients with Wilson disease: rapid detection of mutations by limited sequencing and phenotype-genotype analysis. J Hepatol 2001;35:575-81. [Crossref] [PubMed]
- Todorov T, Savov A, Jelev H, et al. Spectrum of mutations in the Wilson disease gene (ATP7B) in the Bulgarian population. Clin Genet 2005;68:474-6. [Crossref] [PubMed]
- Vrabelova S, Letocha O, Borsky M, et al. Mutation analysis of the ATP7B gene and genotype/phenotype correlation in 227 patients with Wilson disease. Mol Genet Metab 2005;86:277-85. [Crossref] [PubMed]
- Shah AB, Chernov I, Zhang HT, et al. Identification and analysis of mutations in the Wilson disease gene (ATP7B): population frequencies, genotype-phenotype correlation, and functional analyses. Am J Hum Genet 1997;61:317-28. [Crossref] [PubMed]
- Kluska A, Kulecka M, Litwin T, et al. Whole-exome sequencing identifies novel pathogenic variants across the ATP7B gene and some modifiers of Wilson’s disease phenotype. Liver Int 2019;39:177-86. [Crossref] [PubMed]
- Ferenci P. Regional distribution of mutations of the ATP7B gene in patients with Wilson disease: impact on genetic testing. Hum Genet 2006;120:151-9. [Crossref] [PubMed]
- Loudianos G, Dessi V, Lovicu M, et al. Mutation analysis in patients of Mediterranean descent with Wilson disease: identification of 19 novel mutations. J Med Genet 1999;36:833-6. [PubMed]
- Margarit E, Bach V, Gomez D, et al. Mutation analysis of Wilson disease in the Spanish population -- identification of a prevalent substitution and eight novel mutations in the ATP7B gene. Clin Genet 2005;68:61-8. [Crossref] [PubMed]
- Loudianos G, Dessi V, Lovicu M, et al. Molecular characterization of wilson disease in the Sardinian population--evidence of a founder effect. Hum Mutat 1999;14:294-303. [Crossref] [PubMed]
- Garcia-Villarreal L, Daniels S, Shaw SH, et al. High prevalence of the very rare Wilson disease gene mutation Leu708Pro in the Island of Gran Canaria (Canary Islands, Spain): a genetic and clinical study. Hepatology 2000;32:1329-36. [Crossref] [PubMed]
- Figus A, Angius A, Loudianos G, et al. Molecular pathology and haplotype analysis of Wilson disease in Mediterranean populations. Am J Hum Genet 1995;57:1318-24. [PubMed]
- Lo C, Bandmann O. Epidemiology and introduction to the clinical presentation of Wilson disease. Handb Clin Neurol 2017;142:7-17. [Crossref] [PubMed]
- Gu YH, Kodama H, Du SL, et al. Mutation spectrum and polymorphisms in ATP7B identified on direct sequencing of all exons in Chinese Han and Hui ethnic patients with Wilson's disease. Clin Genet 2003;64:479-84. [Crossref] [PubMed]
- Okada T, Shiono Y, Hayashi H, et al. Mutational analysis of ATP7B and genotype-phenotype correlation in Japanese with Wilson's disease. Hum Mutat 2000;15:454-62. [Crossref] [PubMed]
- Yoo HW. Identification of novel mutations and the three most common mutations in the human ATP7B gene of Korean patients with Wilson disease. Genet Med 2002;4:43-48S. [Crossref] [PubMed]
- Jang JH, Lee T, Bang S, et al. Carrier frequency of Wilson’s disease in the Korean population: a DNA-based approach. J Hum Genet 2017;62:815-8. [Crossref] [PubMed]
- Tatsumi Y, Hattori A, Hayashi H, et al. Current state of Wilson disease patients in central Japan. Intern Med 2010;49:809-15. [Crossref] [PubMed]
- Aggarwal A, Chandhok G, Todorov T, et al. Wilson disease mutation pattern with genotype-phenotype correlations from Western India: confirmation of p.C271* as a common Indian mutation and identification of 14 novel mutations. Ann Hum Genet 2013;77:299-307. [Crossref] [PubMed]
- Kumari N, Kumar A, Thapa BR, et al. Characterization of mutation spectrum and identification of novel mutations in ATP7B gene from a cohort of Wilson disease patients: Functional and therapeutic implications. Hum Mutat 2018;39:1926-41. [Crossref] [PubMed]
- Rodriguez-Granillo A, Sedlak E, Wittung-Stafshede P. Stability and ATP binding of the nucleotide-binding domain of the Wilson disease protein: effect of the common H1069Q mutation. J Mol Biol 2008;383:1097-111. [Crossref] [PubMed]
- Huster D, Hoppert M, Lutsenko S, et al. Defective cellular localization of mutant ATP7B in Wilson's disease patients and hepatoma cell lines. Gastroenterology 2003;124:335-45. [Crossref] [PubMed]
- Huster D, Kuhne A, Bhattacharjee A, et al. Diverse functional properties of Wilson disease ATP7B variants. Gastroenterology 2012;142:947-56.e5. [Crossref] [PubMed]
- Dmitriev OY, Bhattacharjee A, Nokhrin S, et al. Difference in stability of the N-domain underlies distinct intracellular properties of the E1064A and H1069Q mutants of copper-transporting ATPase ATP7B. J Biol Chem 2011;286:16355-62. [Crossref] [PubMed]
- Payne AS, Kelly EJ, Gitlin JD. Functional expression of the Wilson disease protein reveals mislocalization and impaired copper-dependent trafficking of the common H1069Q mutation. Proc Natl Acad Sci U S A 1998;95:10854-9. [Crossref] [PubMed]
- Gupta A, Bhattacharjee A, Dmitriev OY, et al. Cellular copper levels determine the phenotype of the Arg875 variant of ATP7B/Wilson disease protein. Proc Natl Acad Sci U S A 2011;108:5390-5. [Crossref] [PubMed]
- Merle U, Weiss KH, Eisenbach C, et al. Truncating mutations in the Wilson disease gene ATP7B are associated with very low serum ceruloplasmin oxidase activity and an early onset of Wilson disease. BMC Gastroenterol 2010;10:8. [Crossref] [PubMed]
- Ferenci P, Stremmel W, Czlonkowska A, et al. Age, sex, but not ATP7B genotype effectively influences the clinical phenotype of Wilson disease. Hepatology 2014;26:49-56.
- Maier-Dobersberger T, Ferenci P, Polli C, et al. Detection of the His1069Gln mutation in Wilson disease by rapid polymerase chain reaction. Ann Intern Med 1997;127:21-6. [Crossref] [PubMed]
- Stapelbroek JM, Bollen CW, van Amstel JK, et al. The H1069Q mutation in ATP7B is associated with late and neurologic presentation in Wilson disease: results of a meta-analysis. J Hepatol 2004;41:758-63. [Crossref] [PubMed]
- Nicastro E, Loudianos G, Zancan L, et al. Genotype-phenotype correlation in Italian children with Wilson's disease. J Hepatol 2009;50:555-61. [Crossref] [PubMed]
- Mihaylova V, Todorov T, Jelev H, et al. Neurological symptoms, genotype-phenotype correlations and ethnic-specific differences in Bulgarian patients with Wilson disease. Neurologist 2012;18:184-9. [Crossref] [PubMed]
- Tarnacka B, Gromadzka G, Rodo M, et al. Frequency of His1069Gln and Gly1267Lys mutations in Polish Wilson’s disease population. Eur J Neurol 2000;7:495-8. [Crossref] [PubMed]
- Chabik G, Litwin T, Czlonkowska A. Concordance rates of Wilson’s disease phenotype among siblings. J Inherit Metab Dis 2014;37:131-5. [Crossref] [PubMed]
- Kegley KM, Sellers MA, Ferber MJ, et al. Fulminant Wilson's disease requiring liver transplantation in one monozygotic twin despite identical genetic mutation. Am J Transplant 2010;10:1325-9. [Crossref] [PubMed]
- Czlonkowska A, Gromadzka G, Chabik G. Monozygotic female twins discordant for phenotype of Wilson's disease. Mov Disord 2009;24:1066-9. [Crossref] [PubMed]
- Senzolo M, Loreno M, Fagiuoli S, et al. Different neurological outcome of liver transplantation for Wilson’s disease in two homozygotic twins. Clin Neurol Neurosurg 2007;109:71-5. [Crossref] [PubMed]
- Yahata S, Yung S, Mandai M, et al. Phenotypes and Chronic Organ Damage May Be Different among Siblings with Wilson's Disease. J Clin Transl Hepatol 2017;5:27-30. [PubMed]
- Cocos R, Sendroiu A, Schipor S, et al. Genotype-phenotype correlations in a mountain population community with high prevalence of Wilson’s disease: genetic and clinical homogeneity. PLoS One 2014;9:e98520. [Crossref] [PubMed]
- Usta J, Wehbeh A, Rida K, et al. Phenotype-genotype correlation in Wilson disease in a large Lebanese family: association of c.2299insC with hepatic and of p. Ala1003Thr with neurologic phenotype. PLoS One 2014;9:e109727. [Crossref] [PubMed]
- Leggio L, Malandrino N, Loudianos G, et al. Analysis of the T1288R mutation of the Wilson disease ATP7B gene in four generations of a family: possible genotype-phenotype correlation with hepatic onset. Dig Dis Sci 2007;52:2570-5. [Crossref] [PubMed]
- Genin E, Feingold J, Clerget-Darpoux F. Identifying modifier genes of monogenic disease: strategies and difficulties. Hum Genet 2008;124:357-68. [Crossref] [PubMed]
- Chuang LM, Tai TY, Wang TR, et al. Esterase D and retinoblastoma gene loci are tightly linked to Wilson's disease in Chinese pedigrees from Taiwan. Hum Genet 1991;87:465-8. [Crossref] [PubMed]
- Poli J, Gasser SM, Papamichos-Chronakis M. The INO80 remodeller in transcription, replication and repair. Philos Trans R Soc Lond B Biol Sci 2017.372. [PubMed]
- Litwin T, Gromadzka G, Czlonkowska A. Apolipoprotein E gene (APOE) genotype in Wilson’s disease: impact on clinical presentation. Parkinsonism Relat Disord 2012;18:367-9. [Crossref] [PubMed]
- Schiefermeier M, Kollegger H, Madl C, et al. The impact of apolipoprotein E genotypes on age at onset of symptoms and phenotypic expression in Wilson's disease. Brain 2000;123:585-90. [Crossref] [PubMed]
- Gu YH, Kodama H, Du SL. Apolipoprotein E genotype analysis in Chinese Han ethnic children with Wilson’s disease, with a concentration on those homozygous for R778L. Brain Dev 2005;27:551-3. [Crossref] [PubMed]
- Kocabay G, Tutuncu Y, Yilmaz H, et al. Impact of apolipoprotein E genotypes on phenotypic expression in Turkish patients with Wilson's disease. Scand J Gastroenterol 2009;44:1270-1. [Crossref] [PubMed]
- Du Q, Luu PL, Stirzaker C, et al. Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics 2015;7:1051-73. [Crossref] [PubMed]
- Jung JS, Jee MK, Cho HT, et al. MBD6 is a direct target of Oct4 and controls the stemness and differentiation of adipose tissue-derived stem cells. Cell Mol Life Sci 2013;70:711-28. [Crossref] [PubMed]
- Xu R, Tao A, Zhang S, et al. Association between patatin-like phospholipase domain containing 3 gene (PNPLA3) polymorphisms and nonalcoholic fatty liver disease: a HuGE review and meta-analysis. Sci Rep 2015;5:9284. [Crossref] [PubMed]
- Vigano M, Valenti L, Lampertico P, et al. Patatin-like phospholipase domain-containing 3 I148M affects liver steatosis in patients with chronic hepatitis B. Hepatology 2013;58:1245-52. [Crossref] [PubMed]
- Trepo E, Pradat P, Potthoff A, et al. Impact of patatin-like phospholipase-3 (rs738409 C>G) polymorphism on fibrosis progression and steatosis in chronic hepatitis C. Hepatology 2011;54:60-9. [Crossref] [PubMed]
- Jenkins CM, Mancuso DJ, Yan W, et al. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem 2004;279:48968-75. [Crossref] [PubMed]
- Pingitore P, Pirazzi C, Mancina RM, et al. Recombinant PNPLA3 protein shows triglyceride hydrolase activity and its I148M mutation results in loss of function. Biochim Biophys Acta 2014;1841:574-80. [Crossref] [PubMed]
- Stattermayer AF, Traussnigg S, Dienes HP, et al. Hepatic steatosis in Wilson disease--Role of copper and PNPLA3 mutations. J Hepatol 2015;63:156-63. [Crossref] [PubMed]
- Gromadzka G, Rudnicka M, Chabik G, et al. Genetic variability in the methylenetetrahydrofolate reductase gene (MTHFR) affects clinical expression of Wilson's disease. J Hepatol 2011;55:913-9. [Crossref] [PubMed]
- Simon I, Schaefer M, Reichert J, et al. Analysis of the human Atox 1 homologue in Wilson patients. World J Gastroenterol 2008;14:2383-7. [Crossref] [PubMed]
- Stuehler B, Reichert J, Stremmel W, et al. Analysis of the human homologue of the canine copper toxicosis gene MURR1 in Wilson disease patients. J Mol Med (Berl) 2004;82:629-34. [Crossref] [PubMed]
- Bost M, Piguet-Lacroix G, Parant F, et al. Molecular analysis of Wilson patients: direct sequencing and MLPA analysis in the ATP7B gene and Atox1 and COMMD1 gene analysis. J Trace Elem Med Biol 2012;26:97-101. [Crossref] [PubMed]
- Weiss KH, Merle U, Schaefer M, et al. Copper toxicosis gene MURR1 is not changed in Wilson disease patients with normal blood ceruloplasmin levels. World J Gastroenterol 2006;12:2239-42. [Crossref] [PubMed]
- Weiss KH, Runz H, Noe B, et al. Genetic analysis of BIRC4/XIAP as a putative modifier gene of Wilson disease. J Inherit Metab Dis 2010;33 Suppl 3:S233-40. [Crossref] [PubMed]
- Sorbello O, Sini M, Civolani A, et al. HFE gene mutations and Wilson's disease in Sardinia. Dig Liver Dis 2010;42:216-9. [Crossref] [PubMed]
- Erhardt A, Hoffmann A, Hefter H, et al. HFE gene mutations and iron metabolism in Wilson's disease. Liver 2002;22:474-8. [Crossref] [PubMed]
- Pfeiffenberger J, Gotthardt DN, Herrmann T, et al. Iron metabolism and the role of HFE gene polymorphisms in Wilson disease. Liver Int 2012;32:165-70. [Crossref] [PubMed]
- Merle U, Stremmel W, Gessner R. Influence of homozygosity for methionine at codon 129 of the human prion gene on the onset of neurological and hepatic symptoms in Wilson disease. Arch Neurol 2006;63:982-5. [Crossref] [PubMed]
- Przybylkowski A, Gromadzka G, Czlonkowska A. Polymorphisms of metal transporter genes DMT1 and ATP7A in Wilson's disease. J Trace Elem Med Biol 2014;28:8-12. [Crossref] [PubMed]
- Barker DJ, Eriksson JG, Forsen T, et al. Fetal origins of adult disease: strength of effects and biological basis. Int J Epidemiol 2002;31:1235-9. [Crossref] [PubMed]
- Barker DJ. Sir Richard Doll Lecture. Developmental origins of chronic disease. Public Health 2012;126:185-9. [Crossref] [PubMed]
- Barker DJ, Thornburg KL. The Obstetric Origins of Health for a Lifetime. Clin Obstet Gynecol 2013;56:511-9. [Crossref] [PubMed]
- Berger SL, Kouzarides T, Shiekhattar R, et al. An operational definition of epigenetics. Genes Dev 2009;23:781-3. [Crossref] [PubMed]
- LaSalle JM, Powell WT, Yasui DH. Epigenetic layers and players underlying neurodevelopment. Trends Neurosci 2013;36:460-70. [Crossref] [PubMed]
- Guo H, Zhu P, Yan L, et al. The DNA methylation landscape of human early embryos. Nature 2014;511:606-10. [Crossref] [PubMed]
- Vogel Ciernia A, LaSalle J. The landscape of DNA methylation amid a perfect storm of autism aetiologies. Nat Rev Neurosci 2016;17:411-23. [Crossref] [PubMed]
- Schroeder DI, Blair JD, Lott P, et al. The human placenta methylome. Proc Natl Acad Sci U S A 2013;110:6037-42. [Crossref] [PubMed]
- Kundaje A, Meuleman W, Ernst J, et al. Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317-30. [Crossref] [PubMed]
- Guo JU, Su Y, Shin JH, et al. Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci 2014;17:215-22. [Crossref] [PubMed]
- Lister R, Mukamel EA, Nery JR, et al. Global epigenomic reconfiguration during mammalian brain development. Science 2013;341:1237905. [Crossref] [PubMed]
- Wang Y, Fischle W, Cheung W, et al. Beyond the double helix: writing and reading the histone code. Novartis Found Symp 2004;259:3-17; discussion 17-21, 163-9. [PubMed]
- Sun CB, Zhang X. Advance in the research on super-enhancer. Yi Chuan 2016;38:1056-68. [PubMed]
- Sun Q, Hao Q, Prasanth KV. Nuclear Long Noncoding RNAs: Key Regulators of Gene Expression. Trends Genet 2018;34:142-57. [Crossref] [PubMed]
- Riordan SM, Williams R. The Wilson’s disease gene and phenotypic diversity. J Hepatol 2001;34:165-71. [Crossref] [PubMed]
- Bethin KE, Cimato TR, Ettinger MJ. Copper binding to mouse liver S-adenosylhomocysteine hydrolase and the effects of copper on its levels. J Biol Chem 1995;270:20703-11. [Crossref] [PubMed]
- Li M, Li Y, Chen J, et al. Copper ions inhibit S-adenosylhomocysteine hydrolase by causing dissociation of NAD+ cofactor. Biochemistry 2007;46:11451-8. [Crossref] [PubMed]
- Medici V, Shibata NM, Kharbanda KK, et al. Wilson's disease: changes in methionine metabolism and inflammation affect global DNA methylation in early liver disease. Hepatology 2013;57:555-65. [Crossref] [PubMed]
- Delgado M, Perez-Miguelsanz J, Garrido F, et al. Early effects of copper accumulation on methionine metabolism. Cell Mol Life Sci 2008;65:2080-90. [Crossref] [PubMed]
- Meng H, Chen G, Gao HM, et al. The emerging nexus of active DNA demethylation and mitochondrial oxidative metabolism in post-mitotic neurons. Int J Mol Sci 2014;15:22604-25. [Crossref] [PubMed]
- Matilainen O, Quiros PM, Auwerx J. Mitochondria and Epigenetics - Crosstalk in Homeostasis and Stress. Trends Cell Biol 2017;27:453-63. [Crossref] [PubMed]
- Yin R, Mo J, Dai J, et al. Nickel(II) Inhibits Tet-Mediated 5-Methylcytosine Oxidation by High Affinity Displacement of the Cofactor Iron(II). ACS Chem Biol 2017;12:1494-8. [Crossref] [PubMed]
- Chervona Y, Arita A, Costa M. Carcinogenic metals and the epigenome: understanding the effect of nickel, arsenic, and chromium. Metallomics 2012;4:619-27. [Crossref] [PubMed]
- Le A, Shibata NM, French SW, et al. Characterization of timed changes in hepatic copper concentrations, methionine metabolism, gene expression, and global DNA methylation in the Jackson toxic milk mouse model of Wilson disease. Int J Mol Sci 2014;15:8004-23. [Crossref] [PubMed]
- Medici V, Kieffer DA, Shibata NM, et al. Wilson Disease: Epigenetic effects of choline supplementation on phenotype and clinical course in a mouse model. Epigenetics 2016;11:804-18. [Crossref] [PubMed]
- Medici V, Shibata NM, Kharbanda KK, et al. Maternal choline modifies fetal liver copper, gene expression, DNA methylation, and neonatal growth in the tx-j mouse model of Wilson disease. Epigenetics 2014;9:286-96. [Crossref] [PubMed]
- Mordaunt CE, Shibata NM, Kieffer DA, et al. Epigenetic changes of the thioredoxin system in the tx-j mouse model and in patients with Wilson disease. Hum Mol Genet 2018;27:3854-69. [PubMed]
- Mordaunt CE, Kieffer DA, Shibata NM, et al. Epigenomic signatures in liver and blood of Wilson disease patients include hypermethylation of liver-specific enhancers. Epigenetics Chromatin 2019;12:10. [Crossref] [PubMed]
- Li X, Feng Z, Tang W, et al. Sex Differences in Clinical Characteristics and Brain MRI Change in Patients With Wilson’s Disease in a Chinese Population. Front Physiol 2018;9:1429. [Crossref] [PubMed]
- Litwin T, Gromadzka G, Czlonkowska A. Gender differences in Wilson's disease. J Neurol Sci 2012;312:31-5. [Crossref] [PubMed]
- Czlonkowska A, Ciesielska A, Gromadzka G, et al. Estrogen and cytokines production - the possible cause of gender differences in neurological diseases. Curr Pharm Des 2005;11:1017-30. [Crossref] [PubMed]
- Garcia-Calzon S, Perfilyev A, de Mello VD, et al. Sex Differences in the Methylome and Transcriptome of the Human Liver and Circulating HDL-Cholesterol Levels. J Clin Endocrinol Metab 2018;103:4395-408. [Crossref] [PubMed]


