
Fusion heals the broken-hearted
Cardiovascular diseases (CVDs) remain one of the most severe conditions facing patients with reported mortality rates of 17.92 million in 2015 (1). Myocardial infractions (MIs), a prevalent form of CVD, trigger localized cardiomyocyte loss after ischemic injury, and the following inflammatory response and deposition of extracellular matrix causes scar formation (2). While significant advances have been made in diagnosing and treating CVDs, there currently are no treatments that can reverse the fibrotic scarring. Fibrosis induces ventricular remodeling, changing the pressure system, and the stressed heart is at greater risk for recurrent MIs and heart failure (2). Whether the scar tissue could be reversed with healthy myocardium has been at the forefront of cardiac research for the past couple decades because the adult mammalian heart is predominantly a quiescent organ, with limited regenerative potential. One particular focus has been directed at whether cardiomyocytes can proliferate in the adult heart after injury to replace lost tissue. Studies in zebrafish (3,4), newts (5), and in neonatal mice (6,7) have revealed that cardiomyocyte proliferation after injury can occur. Another focus was driven by the identification of endogenous, cardiac progenitor cells (CPCs) that sparked intense study and debate over the last decade about the possibility of resident stem cells restoring cardiac function. The possibility for resident progenitors to repopulate lost cardiac tissue could hold the key to restoring cardiac function after injury.
One major breakthrough came in 2001 when it was shown that bone marrow cells (BMCs) could be transplanted into post-MI mice and differentiate into cardiac cell types that reduced the severity of the injury and improve cardiac physiology (8-10). In these experiments, Orlic et al. demonstrated that injecting Lin– c-Kit+ BMCs (Lin, endothelial cell and Kit, stem cell marker) from enhanced green fluorescent protein (EGFP+) mice into the infarcted left ventricle increased the EGFP+ cardiac muscle cells suggesting that c-Kit+ BMCs differentiated into cardiomyocytes (9). They also demonstrated that Lin− c-Kit+ BMCs differentiated into endothelial cells and smooth muscles cells which indicated that these progenitor cells behaved as stem cells since they could give rise to multiple lineages (9). In 2003 Beltrami et al. isolated Lin− c-Kit+ CPCs from adult rat myocardium, cultured under in vitro conditions, and were able to differentiate them into cardiomyocytes, smooth muscle cells, and endothelial cells, however, they were morphologically and functionally immature (11). Moreover, transplanted cardiac-derived Lin− c-Kit+ CPCs into adult rat myocardium post-MI resulted in the appearance of a band of differentiated cardiomyocytes, smooth muscle and endothelial cells within the infarct (11). These initial findings excited the cardiac regeneration field because it showed that resident cells within the myocardium behaved as stem cells and could differentiate into functional cardiac cells lineages to improve recovery after MI.
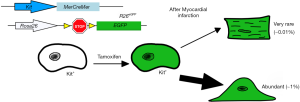
More recently, Ellison et al. expanded on Beltrami’s findings to show that c-Kit+ CPCs were both necessary and sufficient for cardiac regeneration. They administered 5-fluorouracil (an agent that disrupts mitosis) after cardiac injury and found new cardiomyocyte formation and c-Kit+ CPC expansion was absent. In addition, heart failure symptoms in these rats were prevalent (12). Ellison et al. then transplanted GFP− expressing c-Kit+ CPCs into 5-fluorouracil treated rats with MI and found GFP+ cardiomyocytes and vascular cells within the damaged myocardium: suggesting that c-Kit+ CPCs were critical for endogenous regeneration of the heart (12). While these findings increased the interest in manipulating this cell population, other groups attempted to directly test the resident c-Kit+ CPCs contribution to cardiomyocyte differentiation and heart physiology after MI (13-16). van Berlo et al. performed c-Kit+ CPC lineage tracing by developing a mouse line that expressed Cre recombinase and endogenous GFP at the Kit locus and bred these mice into a Rosa26-eGFP line so that all Kit+ cells would express EGFP (Figure 1) (16). They observed minimal expression of EGFP+ cardiomyocytes in uninjured and post-MI hearts (less than 0.03%), but they did observe colocalization of the CD31+ cells and EGFP signal which suggested endothelial cell expansion (16). Interestingly, using the Rosa26-membrane-Tomato-loxP-stop-loxP-membrane-eGFP line (R26mT/mG), van Berlo et al. observed cell-fusion events, and not de novo cardiomyocyte formation as a mechanism of c-Kit+ derived myocyte cells (16). They concluded that this low abundance of observed cardiomyocytes derived from c-Kit+ CPCs suggested that this population was not sufficient to contribute to improving heart function after MI. Similar findings were reported by three other studies (13-15). Sultana et al. further answered these questions by developing multiple knock-in lineages that would express different fluorescent proteins fused with histone H2B protein (15). They crossed mice expressing the knock-in allele c-kitH2B-tdTomato with those containing Nkx2.5H2B-GFP (marking the early cardiogenic transcription factor Nkx2.5) or cTnTH2B-GFP (which labels the mature sarcomeric structures), and found minimal colocalization of these markers within c-Kit+ cells (15). Sultana et al. also observed increased colocalization of c-kit with endothelial protein PECAM and noted c-Kit+ CPCs demonstrated limited differentiation into cardiomyocytes (15). Liu et al. sought to determine the fates of the c-Kit+ CPC-derived cells by reassessing the findings from Ellison and van Berlo, and they employed an instant pulse-chase model where they collected Kit-CreER; Rosa26-RFP hearts at 24 and 48 hours after tamoxifen treatments (14). Using RFP+ cardiac troponin I (Tnni3)+ as markers of c-Kit+ cardiomyocytes, they observed that 58-71% of all cardiomyocytes were labeled by both markers (14). They postulated that these RFP+Tnni3+ cardiomyocytes did not differentiate from Kit+ CPCs, because was unlikely that such a large percentage of myocytes would be generated within 48 hours, and that a population of pre-existing cardiomyocytes expressed c-Kit which could create artifactual findings with basic lineage tracing (14). A more definitive set of experiments were conducted by He et al. (13). They generated a dual-recombinase reporter system to determine if c-Kit+ CPCs differentiated into de novo cardiomyocytes because the traditional, single-recombinase lineage-tracing system often had off-target expression of the fluorescent tracer protein (13). This was achieved by the development of a mouse (Tnni3-Dre × Kit-CreER × IR1) that expressed Dre recombinase under the control of Tnni3, tamoxifen-inducible Cre recombinase controlled by Kit promoter, and an interleaved reporter (IR1) labeling Dre-rox recombination as tdTomato, while ZsGreen is expressed if Cre-loxP recombination occurred (13). He et al. observed a majority of ZsGreen+ endothelial cells but no ZsGreen+ cardiomyocytes in Tnni3-Dre × Kit-CreER × IR1 hearts, while Kit-CreER × IR1 control hearts showed ZsGreen in both cardiomyocytes and non-myocytes (13). These findings confirmed the traditional Cre-recombinase system was not as specific as previously believed, as some cardiomyocytes express Kit, and that a significant portion of the de novo cardiomyocytes previously reported are artifacts of the system, i.e., Kit expression in mature cardiomyocytes. The question still unresolved is whether the observed c-Kit+ derived cardiomyocytes is actually the result of cell-fusion.
In their recent publication, Maliken et al. determined if endogenous CPCs can give rise to de novo cardiomyocytes or fused with resident cardiomyocytes (17). This was addressed by using Kit lineage-specific deletion of the cardiogenic transcription factors Gata4 and Gata6. Gata4 is required for heart tube formation during embryogenesis (18,19) and Gata4/Gata6 double mutants failed to develop a functional heart entirely (20), indicating that Gata4 and Gata6 are necessary genes for cardiac development and cardiomyocyte differentiation. Maliken et al. hypothesized that if CPCs from the Kit lineage were differentiating into de novo cardiomyocytes, then the loss of Gata4 and Gata6 would inhibit this differentiation of CPCs into cardiomyocytes (17). To test this, Maliken et al. generated mouse lines containing tamoxifen-inducible MerCreMer (MCM) under the control of the Kit allele and floxed alleles of Gata4 and Gata6 (Figure 2). In the presence of tamoxifen, Kit+ cells would selectively recombine and disrupt the Gata genes. These lines were also bred to have either the Rosa26-loxP-stop-loxP-eGFP (R26eGFP) to lineage trace all Kit-allele derived cells (17). First, mice were fed tamoxifen after weaning, and were sacrificed after 1-, 2-, or 4-month of treatment to determine if loss of Gata4, Gata6, or both reduced the number of cardiomyocytes derived from the Kit+R26eGFP CPCs. Unexpectedly, loss of Gata4 (Gata4fl/fl) and the double knockout of Gata4/Gata6 (Gata4fl/fl/Gata6fl/fl) increased the number of eGFP+ cardiomyocytes compared to the control mice. These results were surprising because it was previously reported that the deletion of Gata4 and Gata6 blocked cardiomyocyte differentiation and caused acardia (20). To determine if these myocytes were the result of de novo differentiation from the Kit+ CPCs or fusion events, Maliken et al. used KitMCMR26mT/mG mice containing either the floxed allele for Gata4 or Gata4/6 and counted mTomato+/m-eGFP+ cells. This dual reporter method showed an increase in mTomato+/m-eGFP+ cardiomyocytes in Gata4fl/fl and Gata4fl/fl/Gata6fl/fl when compared to controls and indicated the labeled myocytes were from fusion events instead of de novo origins. In the original experiments that showed Kit+ could give rise to multiple lineages in the myocardium, transplanted Lin-Kit+ BMCs were used (8,9,11). In this study, Maliken et al. used BMCs from KitMCM: Gata4fl/fl/Gata6fl/fl mice to determine if other cell lineages were affected. They observed an expansion in CD133+ endothelial progenitors from KitMCM: Gata4fl/fl/Gata6fl/fl BMCs compared to controls which suggests that Gata genes functions in endothelial cells. Previous lineage tracing studies have shown cells derived from KitCre progenitors to be either endothelial cells (CD31+) or leukocytes (CD45+) (16), and Maliken et al. found that CPCs from either KitMCM: Gata4fl/fl or KitMCM: Gata4fl/fl/Gata6fl/fl lineages contained increased levels of CD31+ and CD45+ expression compared to controls (17). These findings further confirmed that Gata4 plays a role in suppressing endothelial cell and leukocyte differentiation since loss of Gata4 allowed for an expansion of these cell populations.
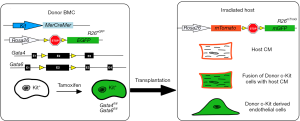
Since the Kit+ CPCs deficient in Gata4 or Gata4/Gata6 displayed increased eGFP+ cardiomyocytes and increased differentiation into endothelial and leukocyte lineages, Maliken et al. next wanted to determine whether the increase in eGFP-expressing myocytes was the result of leukocyte fusion or altered endothelial cell differentiation (17). To understand this phenomenon, they performed two different bone marrow transplantation experiments to determine how loss of Gata4/6 increased eGFP+ myocytes. In the first experiment, they used donor bone marrow from KitMCM:Gata4fl/flGata6fl/fl: R26eGFP (and KitMCMR26eGFP control) mice that underwent 6 weeks of tamoxifen treatment and transplanted this marrow into irradiated recipient R26mT mice (Figure 2) (17). Two months later eGFP+ cardiomyocytes were found in host hearts, and Maliken et al. concluded that these cardiomyocytes were formed from fusion events because they expressed both eGFP and m-Tomato. Maliken et al. also performed flow cytometry on isolated cardiac interstitial cells and found no difference in CD31+ endothelial cell population size which indicated that Gata4fl/fl/6fl/fl bone marrow did not contribute to the expansion in endothelial cells that they originally detected in the KitMCM:Gata4fl/flGata6fl/fl lineage (17). To see if it was the endogenous Kit+ CPCs that were responsible for increased eGFP+ cardiomyocyte presence and CD31+ endothelial cell expansion, they performed a reverse bone marrow experiment. R26mT bone marrow from donor mice were transplanted into irradiated KitMCM:R26eGFP control and KitMCM:Gata4fl/flGata6fl/fl: R26eGFP mice. Mice received 2 months of tamoxifen treatment to inactivate the Gata factors, and analysis of cardiac sections showed increased cardiomyocytes expressing m-Tomato from the donor marrow in KitMCM:Gata4fl/flGata6fl/fl: R26eGFP hearts compared to controls. These findings further support that deletion of Gata4/6 within endogenous Kit+ CPCs is the source of the increase in fusion between native cardiomyocytes and bone marrow-derived leukocytes observed in these hearts.
Since deletion of Gata4/6 within Kit+ CPCs increased progenitor differentiation toward an endothelial lineage, Maliken et al. hypothesized that if they deleted these genes within adult endothelial cells that they would observe weakened vascular permeability since they found greater leukocyte infiltration and fusion to cardiomyocytes. First, they overexpressed vascular endothelial growth factor-A (VEGF-A), a known inducer of endothelial cell permeability, in KitMCM:R26eGFP mice and observed increased eGFP+ cardiomyocytes and leukocyte infiltration. This recapitulated their previous data from the KitMCM:Gata4fl/fl/Gata6fl/fl:R26mT/mG experiment where they lineage traced cardiomyocyte fusion events, suggesting that modulating endothelial leakiness was sufficient to increase the appearance of c-Kit+ derived cardiomyocytes (17). Next, a tamoxifen-inducible Tie2 Cre mouse line (Tie2CreERT2:R26eGFP) was used to specifically disrupt Gata4 in all adult endothelial cells. Tie2CreERT2:R26eGFP controls and Tie2CreERT2:R26eGFP:Gata4fl/fl mice were fed tamoxifen for 2 months and endothelial specific loss of Gata4 displayed a 2-fold increase in eGFP+ adult endothelial cells from increased endothelial cell proliferation. In addition, a shift from CD31+-High expression to CD31+-Low expression was observed, suggesting that Gata4– endothelial cells are less differentiated that negatively altered microvasculature integrity. They also detected increased CD45+ leukocyte presence in Tie2CreERT2:R26eGFP:Gata4fl/fl hearts, which further confirms that vascular permeability has been compromised due to increased leukocyte infiltration (17). This was further confirmed when they bred the KitMCM allele into Tie2CreERT2:R26eGFP:Gata4fl/fl mice to determine if increased cardiomyocyte fusion events would occur when compared to KitMCM or Tie2CreERT2 alone. As predicted, KitMCM: Tie2CreERT2:R26eGFP:Gata4fl/fl hearts showed significantly increased eGFP+ cardiomyocytes and approximately double the number of eGFP+ cells from their KitMCM lineage tracing fusion experiments with loss of Gata4 in both the adult endothelial cells and Kit+ CPCs (17). Finally, R26mT bone marrow transplantation into irradiated Tie2CreERT2:R26eGFP:Gata4fl/fl recipient mice resulted in increased leukocyte-driven cardiomyocyte fusion. These studies demonstrated that the loss of Gata4 in adult endothelial cells allows leukocyte infiltration as vascular permeability is increased in this less differentiated endothelium and that this amplified leukocyte-cardiomyocyte fusion events.
Understanding how endogenous progenitors within the heart respond after injury and whether they can be manipulated into specific cell lineages are critical questions in regenerative medicine. So determining Kit+ CPCs behavior is necessary to advance our current knowledge. Maliken et al. used multiple Cre recombinase lines, under the control of Kit and Tie2 promoters, and show that de novo cardiomyocytes previously thought to differentiate from Kit+ CPCs are the result of leukocyte-fusion and the deletion of Gata4 enhanced the frequency of these events. Cell fusion, specifically bone marrow cell-derived fusion, is a known phenomenon as BMCs were reported to fuse with neurons, Purkinje cells, and cardiomyocytes (21), so the findings here advances our understanding that Gata factors are important in this process. Future studies could explore whether increasing leukocyte to cardiomyocyte fusion can be beneficial to the native cardiomyocytes, especially after myocardial infarction. Orlic, Beltrami, and Ellison saw that endogenous Kit+ CPCs and transplanted Kit+ BMCs improved cardiac physiology (8-12). Thus, leukocyte-fusion may impart a cardioprotective phenotype onto the native cardiomyocytes that allows them to survive within the ischemic environment of the infarct. Elucidating if the leukocyte-fused cardiomyocytes plays an important role post-MI would advance our understanding of how cardiomyocytes can be protected after injury and mend broken hearts.
Acknowledgements
Funding: DA Zuppo and M Tsang are supported by the National Institutes of Health grants 5T32EB001026-14 and R01HL142788, respectively.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Benjamin EJ, Blaha MJ, Chiuve SE, et al. Heart Disease and Stroke Statistics-2017 Update: A Report From the American Heart Association. Circulation 2017;135:e146-603. [Crossref] [PubMed]
- Roger VL, Go AS, Lloyd-Jones DM, et al. Heart disease and stroke statistics--2012 update: a report from the American Heart Association. Circulation 2012;125:e2-220. [PubMed]
- Jopling C, Sleep E, Raya M, et al. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature 2010;464:606-9. [Crossref] [PubMed]
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science 2002;298:2188-90. [Crossref] [PubMed]
- Oberpriller J, Oberpriller JC. Mitosis in adult newt ventricle. J Cell Biol 1971;49:560-3. [Crossref] [PubMed]
- Darehzereshki A, Rubin N, Gamba L, et al. Differential regenerative capacity of neonatal mouse hearts after cryoinjury. Dev Biol 2015;399:91-9. [Crossref] [PubMed]
- Porrello ER, Mahmoud AI, Simpson E, et al. Transient regenerative potential of the neonatal mouse heart. Science 2011;331:1078-80. [Crossref] [PubMed]
- Orlic D, Kajstura J, Chimenti S, et al. Bone marrow cells regenerate infarcted myocardium. Nature 2001;410:701-5. [Crossref] [PubMed]
- Orlic D, Kajstura J, Chimenti S, et al. Transplanted adult bone marrow cells repair myocardial infarcts in mice. Ann N Y Acad Sci 2001;938:221-9; discussion 229-30. [Crossref] [PubMed]
- Orlic D, Kajstura J, Chimenti S, et al. Mobilized bone marrow cells repair the infarcted heart, improving function and survival. Proc Natl Acad Sci U S A 2001;98:10344-9. [Crossref] [PubMed]
- Beltrami AP, Barlucchi L, Torella D, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 2003;114:763-76. [Crossref] [PubMed]
- Ellison GM, Vicinanza C, Smith AJ, et al. Adult c-kit(pos) cardiac stem cells are necessary and sufficient for functional cardiac regeneration and repair. Cell 2013;154:827-42. [Crossref] [PubMed]
- He L, Li Y, Li Y, et al. Enhancing the precision of genetic lineage tracing using dual recombinases. Nat Med 2017;23:1488-98. [Crossref] [PubMed]
- Liu Q, Yang R, Huang X, et al. Genetic lineage tracing identifies in situ Kit-expressing cardiomyocytes. Cell Res 2016;26:119-30. [Crossref] [PubMed]
- Sultana N, Zhang L, Yan J, et al. Resident c-kit(+) cells in the heart are not cardiac stem cells. Nat Commun 2015;6:8701. [Crossref] [PubMed]
- van Berlo JH, Kanisicak O, Maillet M, et al. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature 2014;509:337-41. [Crossref] [PubMed]
- Maliken BD, Kanisicak O, Karch J, et al. Gata4-Dependent Differentiation of c-Kit(+) Derived Endothelial Cells Underlies Artefactual Cardiomyocyte Regeneration in the Heart. Circulation 2018;138:1012-24. [Crossref] [PubMed]
- Kuo CT, Morrisey EE, Anandappa R, et al. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev 1997;11:1048-60. [Crossref] [PubMed]
- Molkentin JD, Lin Q, Duncan SA, et al. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev 1997;11:1061-72. [Crossref] [PubMed]
- Zhao R, Watt AJ, Battle MA, et al. Loss of both GATA4 and GATA6 blocks cardiac myocyte differentiation and results in acardia in mice. Developmental biology 2008;317:614-9. [Crossref] [PubMed]
- Alvarez-Dolado M, Pardal R, Garcia-Verdugo JM, et al. Fusion of bone-marrow-derived cells with Purkinje neurons, cardiomyocytes and hepatocytes. Nature 2003;425:968. [Crossref] [PubMed]


