Hypertension genomics and cardiovascular prevention
Introduction
Hypertension continues to be a major cause of worldwide mortality and morbidity (1), with genomics proposed to have the potential to assist in reducing the overall burden of cardiovascular events (2). The role of genomics has stretched from the initial discovery of monogenetic diseases with large effects (3), to large-population genome-wide association studies (GWAS) detecting common genetic variations with modest effect sizes. The recent publication of the largest cardiovascular genetic association study to date, with over 1 million participants, demonstrated the total number of genetic signals associated with hypertension surpassing 1000, at 901 genetic loci (4). Each subsequent GWAS iteration continues to increase our understanding of the genetic architecture of hypertension and cardiovascular disease. Here, we review the promise of translating genomics into clinical application through potential novel treatment options, risk scores, gene-environment interactions, or pharmacogenetics.
Blood pressure genomics
The vast information provided by large GWAS has resulted in greater understanding of the polygenic nature of blood pressure regulation, where numerous single nucleotide polymorphisms (SNPs) act additively to impact on cardiovascular disease. However, the translation into establishing the underlying genetic mechanism remains difficult. The key barrier is that the causal variant might not be readily identified by the lead GWAS SNP. Instead, the lead SNP indicates a chromosomal region where the causal gene may typically reside within a 500 kb genetic window, with other SNPs in high linkage disequilibrium (LD) (5). This window however only serves as broad guidance, with the increasing understanding of the 3-dimensional configuration of DNA as being important to genetic function, particularly with the discovery of various genomic regions with high levels of local chromatin interactions implicating longer-range interactions (6). Chromatin interaction Hi-C studies aim to identify long-range target genes of non-coding SNPs, and the recent blood pressure GWAS has identified up to 484 long-range interactions, for example between the SLC30A10 locus and the TGFB2 gene being 1.2 Mb apart (4). To compound this complexity, the potential for trans-acting regulatory elements (7) makes identifying the functional variant difficult to pinpoint. Furthermore, a significant proportion of GWAS-significant SNPs is intergenic or near genes without any obvious connection to cardiovascular disease.
To date, there has been some success in exploring the functional impact of these genetic variants. Perhaps the best example remains the UMOD (uromodulin) gene where the 5’ SNP rs13333226 was identified as associated with hypertension in an early GWAS (8). Subsequently, UMOD-deficient mice demonstrated increased sequestration of the loop diuretic target sodium-potassium-chloride co-transporter 2 (NKCC2) in subapical vesicles together with reduced phosphorylation, both combining with resultant reduced co-transporter activity (9). Mimicking the effect of loop diuretics, this resulted in increased natriuresis and a 20 mmHg lower blood pressure in knockout mice. The BP difference was exacerbated with salt-loading, where the knockout mice were resistant to its hypertensive effects (10). Conversely, the blood pressure of UMOD transgenic mice were salt-sensitive (11).
More recent successes include exploring the genetic function of blood pressure loci, including NPR3 (12), SLC4A7 (13) and SLC39A8 (14). The BP-raising allele at the NPR3 (natriuretic peptide receptor C) locus was associated with altered chromatin interactions, increased NPR3 expression, linked to increased vascular smooth muscle cell proliferation, angiotensin II-induced calcium flux and cell contraction (12). Vascular smooth muscle has also been shown to be relevant to the SLC4A7 (electroneutral sodium-bicarbonate cotransporter 1) locus. The BP-raising allele was associated altered chromatin interactions, increased gene expression, elevated steady-state intracellular pH and accelerated recovery from intracellular acidosis, all independent of the missense polymorphism resulting in the amino acid substitution Glu326Lys (13). Vascular endothelial cells appear to have a greater influence with the SLC39A8 locus, encoding ZIP8, a heavy metal ion transporter. The blood pressure polymorphism is associated with an Ala391Thr variation where blood pressure raising variant Ala391 demonstrated a higher propensity to cadmium accumulation, increased ERK2 phosphorylation, NFkB activation, and reduced vascular endothelial cell viability (14).
With the increased number of genetic loci identified, experimental exploration of each individual locus becomes unfeasible. Despite these complexities, there is still hope that future therapeutic targets could be identified within these genetic loci. There has been a rapid expansion of bioinformatics tools that can assist in prioritising areas to optimise the use of resources to identify new therapeutic options. The process of investigating genetic variants can be assisted by in silico analysis, indicating loci with eQTLs in tissues of interest (e.g., GTEx, www.gtexportal.org), DNase I hypersensitivity sites (e.g., DeepSEA, http://deepsea.princeton.edu/), as well as a handful of non-synonymous polymorphisms that have been predicted to be damaging (e.g., SIFT, http://sift.jcvi.org/; and PolyPhen, http://genetics.bwh.harvard.edu/pph2/). Druggability analyses have provided new genetic support for known anti-hypertensive targets (4), with genetic loci including targets of established antihypertensive medications such as SLC12A2 (loop diuretics), CACNA1C and CACNB4 (calcium channel blockers), within the pathway itself such as NOS3 (nitric oxide donors), targets under investigation EDN1 (endothelin 1), NPR1 and NPR3 (natriuretic peptide analogues), and ENPEP (aminopeptidase A inhibitors), or drugs with known antihypertensive effects that could allow for repurposing such as sodium-glucose co-transporter 2 inhibitors for diabetes mellitus (SLC5A1). This demonstrates, as a proof of concept, the capabilities of genetic studies. In other words, the ability to confirm genetic associations for genes that are the targets of current anti-hypertensive drug targets, then it provides hope that some of the other newly discovered genes for blood pressure may also have the potential to lead to new and improved drugs for hypertension in the future.
In silico functional analyses on gene expression have also highlighted the enrichment of genes relating most strongly to the vasculature, and to a lesser extent, adrenal and adipose tissue. From pathway analyses, there was also an enrichment of signals within the transforming growth factor-β (TGFβ) pathway (4), which is a pathway known to influence renal sodium handling and ventricular remodelling. Furthermore, plasma TGFβ levels have been correlated with hypertension (15). The genes implicated include the growth factor itself (TGFB2), its receptors (TRFBR2 and TGFBR3), downstream signalling proteins such as the activin A receptor type 1C (ACVR1C) and bone morphogenetic protein 2 (BMP2), and transcription factors important in TGFβ signalling, such as Kruppel-like family 14 (KLF14) which regulates expression of TGFβ receptors (16). This might suggest members of this pathway as future novel therapeutic targets.
In recent years, there has also been an increased interest in epigenomic-wide association studies (EWAS). Analogous to GWAS utilising SNPs, these studies utilise quantifiable epigenetic marks, typically DNA methylation, to identify loci that can discriminate between cases and controls (17). EWAS, combined with gene expression analyses have identified six genes (TSPAN2, SLC7A11, UNC93B1, CPT1A, PTMS, and LPCAT3) with mutual associations between methylation, gene expression, and blood pressure. These genes have hitherto not been implicated in the pathogenesis of hypertension with GWAS, indicating a distinct and cumulative gain of knowledge with from this complementary methodology (18). Like its genomic counterpart, EWAS too has limitations. Epigenetic variations may arise as either a cause or a consequence of disease, and can be difficult to differentiate without the use of expensive and time-consuming longitudinal cohort studies. In addition, samples currently utilised for EWAS for are almost invariably blood, which may not reflect the unique epigenetic signature of the tissue of interest.
Genomics of blood pressure and other cardiovascular risk factors
In terms of prioritising research for potential future therapies, it may be reasonable to consider genetic loci that are signals across other cardiovascular risk factors in addition to hypertension, which often co-exist (Table 1) (4,19-24). These signals of interest include B-Cell CLL/lymphoma 2 (BCL2), carbamoyl-phosphate synthase 1 (CPS1) and melatonin receptor 1B (MTNR1B). The rs79598313/KDF1 (keratinocyte differentiation factor 1) locus is more complex as the genes within this LD block also includes AT-Rich Interaction domain 1A (ARID1A), nuclear distribution c, dynein complex regulator (NUDC) and zinc finger DHHC-type containing 18 (ZDHHC18), where little is known of these gene products in relation to pathogenesis of hypertension. As there is a large body of evidence already on APOE in cardiovascular research, this locus is not reviewed in detail here.

Full table
BCL2 is a known inhibitor of apoptosis (25), and is positioned within the angiotensin II-induced endothelial apoptosis pathway (26). However, it is unclear whether modulating BCL2 function has an impact on endothelial survival or function. BCL2 also has a role in a range of tissues, and its upregulation in numerous tumours has made it a potential cancer therapeutic target (27). With this, there is potential for off-target effects and may give reason to pause if considering BCL2 as an anti-hypertensive/diabetic therapy.
Carbamoyl-phosphate synthase 1 (CPS1) catalyses the rate-limiting step in the urea cycle and L-citrulline production. Vascular endothelial cells synthesize endogenous L-arginine by recycling L-citrulline, the by-product of nitric oxide synthesis, using components of the urea cycle, potentially linking nitric oxide production and the urea cycle. There is already some support for CPS1 as a regulator of vascular tone, where the naturally occurring T1405N variation has been observed to influence forearm blood flow responses to bradykinin and nitroprusside, and levels of nitric oxide metabolites (28). There is less known about the biological link between CPS1 and HDL cholesterol, except that there the proteomic changes in murine adipose tissue following a high fat diet were primarily within the urea cycle, including CPS1 (29). There may be however concerns for off-target effects when modulating CPS1 activity. Expression data from GTEx suggests that CPS1 is predominantly expressed in the liver. Furthermore, CPS1 deficiency is a rare autosomal recessive inherited disease resulting in severe hyperammonaemia and protein intolerance (30).
MTNR1B encodes a high affinity receptor for melatonin (31), and appears to influence 24-hour non-rapid eye movement sleep (32). There is some epidemiological support for the influence of melatonin on circadian blood pressure (33), and melatonin reducing nocturnal BP in a small clinical trial (34). This discovery of a melatonin receptor as a genetic signal for blood pressure may provide further impetus to revisit melatonin as a therapeutic target. There is a larger body of evidence for MTNR1B in type II diabetes mellitus in terms of genetic/genomic analyses, clinical/epidemiology data, functional analyses of genetic polymorphisms, in vitro and animal model, where there are still controversies on the potential relationship (35).
Genomics of blood pressure and other cardiovascular endpoints
An alternative aid prioritising genetic loci to undergo functional assessment would consider those that overlap with the genetic signals of cardiovascular endpoints. This approach is however limited by the heterogeneous pathophysiology of cardiovascular endpoints, particularly with heart failure, stroke and chronic renal disease . Furthermore, there is notable variation in classification of phenotypes within each cardiovascular endpoint (e.g., chronic kidney disease has also been investigated under phenotype classifications of end-stage renal failure, creatinine and kidney function decline). Another limitation is the lower prevalence of heart failure and stroke, and subsequently, a limited number of large GWAS (Table 2) (24,36-41).
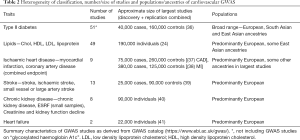
Full table
Signals of association that overlap between blood pressure and coronary artery disease GWAS include apolipoprotein E (APOE), endothelin receptor type A (EDNRA) and SWAP Switching B-Cell Complex Subunit 70 (SWAP70). Overlapping GWAS signals for blood pressure and renal function include vascular endothelial growth factor A (VEGFA) and the aforementioned carbamoyl-phosphate synthase 1 (CPS1) (Table 3) (4,37,38,40,42,43). The impact of apolipoprotein E and endothelin on cardiovascular disease has been well described and not discussed further in this review.

Full table
SWAP70 belongs to a family of proteins involved in an array of processes that control autoimmune phenotypes which spontaneously develop in their absence (44), where one of its homologues is associated with the development of systemic lupus erythematosus (SLE) (45). Autoimmune diseases are increasingly recognised as a risk factor for cardiovascular disease, with the latest cardiovascular risk calculator, QRISK3, having added SLE into the latest iteration (46). Most of the studies on SWAP70 thus far centre on immune cells. However, in context of Kaposi sarcomas, SWAP70 was found to be crucial for in vitro endothelial tube formation and endothelial sprouting (47). The relevance of this finding to endothelial cells in blood pressure regulation is unclear.
The presence of VEGFA on this list is not surprising due to the side effect profile of anti-VEGF cancer therapies with increased risk of hypertension, proteinuria and myocardial infarctions (48,49). The proposed mechanism would be via both vascular and renal endothelial cells, and podocytes. Reduced VEGF activity in the vascular endothelium could lead to vascular rarefaction and reduced nitric oxide availability. Within the kidneys, there could also be downregulation of tight junctions, resulting in proteinuria (48). The importance and opposing effect of VEGF in cancer pathways suggests that it is less likely to be a successful candidate target for cardiovascular disease.
It is also notable that the ABO gene (α-1,3-N-acetylgalactosaminyltransferase and α-1,3-galactosyltransferase), most commonly known for its influence on the ABO blood group, is a signal across multiple traits, including blood pressure (4), glycated haemoglobin A1c (50), hypercholesterolaemia (51), and ischaemic heart disease (52), but with various SNPs not in high LD. The ABO blood group has long been an established risk factor for arterial thrombosis (53), and a recent meta-analysis, the clinical phenotype of ABO blood group itself is a risk factor for coronary artery disease, where blood group A carried the highest risk, and lowest risk with blood group O (54). This may be in part related to the presence of N-linked oligosaccharide side chains on von Willebrand factor (vWF) molecules that contain A and B blood group antigens which in turn decreases von Willebrand factor clearance (55). Individuals with non-O (A, B, or AB) blood groups have 25% higher vWF levels than individuals with blood group O (56). There may also be a role for angiotensin converting enzyme in this relationship between ABO and cardiovascular disease. Both the ABO genotype (57,58) and blood group phenotype (59) are associated with angiotensin-converting enzyme activity. This may in part provide the biological link with hypertension. The relationship between the ABO locus and type II diabetes and hypercholesterolaemia may lie with the link with pancreatic lipase levels varying with ABO genotype at GWAS-significance levels (57), but this hypothesis still requires further study. Overall, this may suggest that α-1,3-N-acetylgalactosaminyltransferase or α-1,3-galactosyltransferase may be a possible therapeutic target, spanning multiple cardiovascular comorbidities.
Genetic risk scores
Outside of generating new pharmacological targets, a different route that genomics can potentially influence clinical care is via augmenting the predictive value of clinical risk scores. Clinical risk scores have long been in use to estimate the actual risk of developing a disease of a defined population, and that the absolute risk of cardiovascular disease is influenced by the combination of risk factors (60,61). Likewise, even though each blood pressure-associated variant only has a small effect individually, a genetic risk score (GRS) can consider the larger aggregated effects of all combined variants. Clinical interventions (both pharmacological and lifestyle) can be effective in delaying the disease progression from prehypertension to hypertension, and the development of cardiovascular events, but also carries the risk of adverse events, as well as financial and opportunity costs. With this, improvements in prediction models to stratify patient populations according to risk would allow a precision medicines strategy to prevent future cardiovascular disease. Genetic risk scores aim to add to clinical risk scores to enhance its predictive value.
The combination of the all known BP variants across 901 loci was associated with a 10.4 mmHg higher SBP, and an over three-fold sex-adjusted higher risk of hypertension (OR 3.34), and odds ratio of incident cardiovascular events of 1.52 comparing top-bottom GRS deciles (4). This predictive ability of GRS highlights the potential to influence clinical management by improved risk stratification. However, this does not assess the utility of GRS in addition to current clinical risk scores. A study using only 22 blood pressure variants as part of a genetic risk score improved discrimination for incident hypertension on top of clinical risk factors, but only modestly (C-index change =0.3–0.5%) (62). While this only showed a modest change, it only utilised a small fraction of known loci and there is potential to improve the discriminatory power by using all the 901 known loci.
To assess the impact of genetics and exposure to lifestyle factors on blood pressure, a genetic risk score composed of 314 blood pressure loci was assessed together with a healthy lifestyle score (BMI, sedentary hours, alcohol intake, meat intake, urinary sodium excretion, fruit and vegetable intake, fish intake and smoking status). For all genetic risk score tertiles, a healthier lifestyle score is associated with lower blood pressure and improved outcomes (63). A separate study focused on the impact of genetic influence on salt-sensitivity with participants undertaking specific dietary interventions of low-sodium, high-sodium and high-sodium/potassium-supplemented diets. Higher GRS conferred larger rises in blood pressure when exposed to a high-sodium diet, but a smaller blood pressure fall with a low-sodium diet. However, the overall influence of the GRS groups is far smaller than that of the dietary interventions itself (64). Taken together, this emphasises that lifestyle management should be for the whole population, rather than targeted using genetic information.
There may however be utility for GRS within a “precision medicine” approach. An example could be seen in studies where GRS improved clinical risk score C-statistics of predictive coronary artery disease in the region of 0.4% to 1% (65,66), utilising between 20 to 50 SNPs in these GRS. Importantly, this small SNP panel resulted in net reclassification by 5% to 9% (66). The improved reclassification to influence the decision to initiate treatment with statins has numbers-needed-to-treat (NNTs) in the ranging between 20 and 60 s, being lower for those with the highest genetic risk (66,67). GRS has also been proposed as a potential motivator in adherence to lifelong pharmacological therapies and behavioural changes, particularly in use for counselling patients with those at higher risk categories. Adding GRS to standard-of-care in counselling patients with coronary artery disease may produce some benefits in changing behaviours. In randomised controlled trials, the additional knowledge of their GRS resulted in modest weight loss and increased physical activity (68), and improvements in LDL-cholesterol (69), but requires further evaluation particularly to consider whether it impacts on clinical end-points. This added GRS-based counselling could be important in translating into management of patients with resistant hypertension, where non-adherence to medications is known to be high (70).
There is however other barriers before GRS reach clinical practice. While there may be some clinical benefit from GRS, it is well worth considering the cost implications of genotyping arrays, particularly as it would involve a large screening population, and in view of the potentially large NNTs. There should also be consideration of the potential ethical impact of such risk scoring and the potential to impact on day-to-day lives of the general population receiving genetic risk score results. At the time of this review, several countries have chosen not to adopt laws to specifically prohibit access to genetic data for purposes of life insurance. Several other countries have either adopted laws or developed voluntary moratoria with the industry to prevent this access (71). The impact would be regional, particularly as the perceptions and importance of life insurance varies from country-to-country, and within countries itself. There would also be the cost implications of providing the necessary counselling that should be provided together with such results.
Hypertension gene-environment (GxE) interaction studies
Another method of elucidating the impact of genetics on the pathogenesis of hypertension is through gene-environment (GxE) interaction studies. The model of GxE studies is based on the hypothesis that individuals may be more vulnerable to the negative effects of environmental adversity, or alternatively, more responsive to positive environmental experiences. GxE studies can also reveal further blood pressure-associated loci that can only be detected via an adjustment of, or interaction with, environmental exposure.
A productive region of GxE research thus far is with salt-sensitivity, where the GenSalt consortium identified up to 9 genetic loci interacting with dietary salt intake to influence blood pressure (72). Potential therapeutic targets that may arise from these findings is from the CASP4 (Caspase 4) and MAP kinase interacting serine/threonine kinase 1 (MNK1) loci. Caspase 4 is a protein in the cysteine-aspartic acid protease family that plays an import role in inflammation and innate immunity. The loss of proximal tubules and renal injury in nephropathic cystinosis appears to be associated with overexpression of the CASP4 gene (73). In view of the importance of renal sodium filtration and reabsorption, further studies on the potential role of CASP4 in salt-sensitivity may be warranted. MNK1 gene functions as a Ser/Thr protein kinase that interacts with ERK1 and p38 mitogen-activated protein kinase (MAPK) (74), a pathway that is involved in BP regulation through norepinephrine and angiotensin II (75). Its pathophysiological role in salt-sensitivity is unclear, but due to its position in a known blood pressure regulating pathway, may be an area of fruitful investigation.
Other gene-environment interactions for blood pressure identified so far include 15 genetic loci identified to interact with cigarette smoking to influence blood pressure (76). Additionally, solute carrier family 16 member 9 (SLC16A9, also known as monocarboxylic acid transporter 9) interacting with alcohol consumption (77), and transmembrane protein 182 (TMEM182) with body-mass index (78), where the biological relevance for both findings are currently uncertain.
Hypertension pharmacogenetics
Worldwide, optimal blood pressure management is achieved in fewer than 40% of those treated, despite the availability of a considerable number of drugs from different pharmacological classes (79). Although there are numerous contributing factors for this, one is the degree of genetic-based inter-subject variation in response to different pharmacological classes. Pharmacogenomics have been proposed to have the potential to identify genetic signals that could predict therapeutic effect or adverse outcomes for different drug classes (80). Currently, decisions on antihypertensive drug therapy selection may be based on age and ancestry (81-83), which in turn acts as a surrogate for plasma renin activity (84). It remains that any use of pharmacogenetics requires an increase in predictive value in addition to current clinical stratification.
While candidate gene studies are not particularly common in the current era of genome-wide studies, one of the strongest evidence for pharmacogenetics relates to the genetic locus at ADRB1 (β1-adrenoceptor) and the blood pressure response to β-blockers (85), which has also since been shown to impact on heart failure outcomes (86). As only very few genetic variants yielded pharmacogenetics effects individually, risk score models combining the effects of multiple polymorphisms have been investigated. For example, within the Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) study, using a risk score including SNPs within the FGF5, CHIC2, MOV10, and HFE genes, reveals a potential difference in response to β-blockers in the magnitude of 14/20 mmHg (P=3.3×10−6 for SBP; P=1.6×10−6 for DBP) comparing carriers of one vs. six risk alleles (87).
Another candidate gene study was based on the knowledge renal tubular expression of epithelial Na+-channel (ENaC) which is known to be influenced by a functional neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase (NEDD4L) polymorphism. A subset of the NORDIL (Nordic Diltiazem) trial revealed a pharmacogenetics effect at rs4149601, where carriers of the variant associated with higher ENaC expression had a greater reduction in blood pressure for patients taking β-blocker or diuretic monotherapy but not the calcium channel blocker diltiazem (88). The genetic effect (around 4.5/1.5 mmHg) is, however, modest compared the overall therapeutic effect of these medications (around 15–19/14–15 mmHg), and there is no clear indication that knowledge of the genotype can influence drug choice in a clinically significant manner.
The exploration of pharmacogenetics of antihypertensive therapies has since reached the GWAS era. Thus far, pharmacogenetics influences on the efficacy of a diuretic (hydrochlorothiazide) have been shown with variants at the LYZ-FRS2-YEATS4 (Bonferroni corrected P=0.024) (89), PRKCA (P=3.3×10−8) (90) loci with allelic effects within the region of 3–8/2–4 mmHg, with the GNAS-EDN3 locus approaching genome-wide significance (P=5.5×10−8) (90). Both protein kinase Cα (PRKCA) and GNAS Complex Locus and endothelin 3, respectively (GNAS-EDN3) loci encode proteins involved in calcium signalling and vascular smooth muscle contraction, but the potential biological relevance of LYZ (lysozyme), FRS2 (fibroblast growth factor receptor substrate 2) or YEATS4 (YEATS domain containing 4) is currently unclear. There has also been a reported pharmacogenetic impact of the SLC25A31 rs201279313 deletion genotype influencing blood pressure response to β-blockers in a study limited to African Americans (P=2.5×10−8), with the LRRC15 locus approaching genome-wide significance (P=7.2×10−8). The relevance of both these genes in the blood pressure regulation or the pharmacokinetics/pharmacodynamics of β-blockers is unclear (91). However, as these studies only assessed the response to one drug class, it would not assist in decision making between different the use of different therapeutic drug classes. A following Genetic Epidemiology of Responses to Antihypertensives (GERA) study aim to identify SNPs with pharmacogenetic effects exhibiting opposite direction associations with BP response between diuretic and angiotensin II receptor blocker treatments, but the results were not replicated in an independent study, with none of the SNPs attaining genome-wide significance (92). A pharmacogenetics GWAS study randomly allocating patients to bisoprolol, losartan, HCTZ or amlodipine as monotherapy in a cross-over design initially demonstrated three SNPs (at the ACY3 gene) were significantly associated to BP response to bisoprolol, but none were successfully replicated (93).
Adverse drug reaction have a role in medicines non-adherence, which in turn contributes to suboptimal blood pressure management. Therefore, the ability to predict the likelihood of adverse drug events may be useful. The ACE inhibitor-induced cough is common, and often necessitates a change in drug class to an angiotensin receptor blocker. This adverse effect has been shown in variations in ABO haplotype (94,95), SLCO1B1 (96), KCNIP4 (97), BDKRB2 (94), NK2R (98) and the ACE insertion/deletion variant (99), although these variants were not detected in a recent pharmacogenetics GWAS (100). Thiazide-induced hyponatraemia is also common and can have severe consequences. This adverse reaction has been recently shown to be associated with 14 genetic regions, with further testing indicating a non-synonymous variation of solute carrier organic anion transporter family member 2a1 (SLCO2A1), also known as prostaglandin transporter, also showing a phenotype of intravascular volume expansion, free water reabsorption, urinary prostaglandin E2 excretion, and reduced excretion of serum chloride and antidiuretic hormone (101). Pharmacogenetic GWAS have also identified up to 6 genetic signals for hydrochlorothiazide-induced hyperuricaemia (102).
Despite these advances, pharmacogenetics in hypertension is still far from clinical practice, and requires comparison against successes elsewhere. Pharmacogenetics of predicting adverse drug events has had success with HLA-B*5701 screening for hypersensitivity to the anti-HIV-therapy, abacavir, where there is high predictive value and the ability to prevent a severe, life-threatening reaction (103). This has perhaps set an exceedingly high standard of impact that a pharmacogenetic test for adverse drug reactions should achieve. The antihypertensive pharmacogenetic studies thus far have only provided some mechanistical insights, but without the necessary predictive values and to influence the choice of antihypertensive drugs.
Our current understanding of pharmacogenetics is often complicated by datasets that includes polypharmacy (including non-cardiovascular medications), numerous drugs and dosing ranges within each antihypertensive drug class, resulting in multiple confounding factors. To minimise these confounders, most pharmacogenetic GWAS so far have used subsets of randomized controlled trials comparing different classes of antihypertensive drugs, for which consenting subjects have subsequently been genotyped, for example the GenHAT study as a subset of the ALLHAT study (104). Furthermore, studies in pharmacogenetics still lag behind the large sample sizes of GWAS. In the context of clinically-predetermined guidelines for first-line (and even second- and third-line) drug choices (83), it may be difficult to obtain sufficient new data that would be able to compare pharmacogenetic effects. With this, the International Consortium for Antihypertensive Pharmacogenomics Studies (ICAPS) was formed in 2012 to increase the opportunities to discover and replicate genetic signatures of many different phenotypes related to antihypertensive treatment response. To date, no signals reach genome-wide significance for influencing the impact of diuretics on blood pressure (105). Alternatively, consortia such as the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium (http://www.chargeconsortium.com) have included observational studies within pharmacogenetics analyses, using longitudinal cohorts containing accurate medical records for drug exposure. Similarly, this has so far been unable to identify any genome-wide significant interactions from four antihypertensive therapy meta-analyses for cardiovascular outcomes (ACE inhibitor/angiotensin receptor blockers, β-blockers, calcium channel blockers or diuretics) (106).
While initial evidence being limited, there is still hope for pharmacogenetic studies to expand in sample sizes, potentially identifying genetic variants with contrasting associations and unique effects to different classes of antihypertensive drugs. There are still many barriers before being able to reach clinical application, which would also need to require the consideration of cost-effectiveness, particularly in the presence of only marginal gains.
Summary
In light of the largest cardiovascular GWAS to date, this review considered whether the knowledge of hypertension genomics has made an impact on clinical practice, and how it may do so in the future. Of the numerous loci and genes implicated in the pathophysiology of hypertension, UMOD has shown promise as a new pharmacological target, and there is a strong enrichment of targets within the TGF-β pathway. In the future, there may be a role for combining GWAS of other comorbidities or cardiovascular endpoints to identify targets that may have a dual effect, where CPS1, MTNR1B and ABO may be the best options highlighted. The development of large DNA biobanks with dense phenotypic information would also allow future studies in the form of phenome-wide association studies (PheWAS), where well-curated electronic health records would allow investigators to use a variety of input functions such as single/multiple SNPs, drug exposure or predicted gene expression to probe broader phenotypes (107). GxE studies may also be important in this aspect, with CASP4 appearing as an interesting candidate gene for salt-sensitivity. Pharmacogenetic and genetic risk score studies have also reveal some exciting mechanistical insights, but clinical application currently remains a distant prospect. With an expansion in the sample sizes of studies, the combination of multiple genetic signals may be sufficient to achieve clinical significance in the future. Should the technological costs of assessing panels of genetic variants decrease, there may still be cost-effectiveness in these measures.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Forouzanfar MH, Liu P, Roth GA, et al. Global Burden of Hypertension and Systolic Blood Pressure of at Least 110 to 115 mm Hg, 1990-2015. JAMA 2017;317:165-82. [Crossref] [PubMed]
- Pratt RE, Dzau VJ. Genomics and hypertension: concepts, potentials, and opportunities. Hypertension 1999;33:238-47. [Crossref] [PubMed]
- Lifton RP. Molecular genetics of human blood pressure variation. Science 1996;272:676-80. [Crossref] [PubMed]
- Evangelou E, Warren HR, Mosen-Ansorena D, et al. Genetic analysis of over one million people identifies 535 novel loci for blood pressure. bioRxiv 2017. Available online: https://www.biorxiv.org/content/early/2017/10/11/198234
- Munroe PB, Barnes MR, Caulfield MJ. Advances in blood pressure genomics. Circ Res 2013;112:1365-79. [Crossref] [PubMed]
- Schmitt AD, Hu M, Jung I, et al. A Compendium of Chromatin Contact Maps Reveals Spatially Active Regions in the Human Genome. Cell Rep 2016;17:2042-59. [Crossref] [PubMed]
- Bouwman BA, de Laat W. Getting the genome in shape: the formation of loops, domains and compartments. Genome Biol 2015;16:154. [Crossref] [PubMed]
- Padmanabhan S, Melander O, Johnson T, et al. Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet 2010;6. [Crossref] [PubMed]
- Mutig K, Kahl T, Saritas T, et al. Activation of the bumetanide-sensitive Na+,K+,2Cl- cotransporter (NKCC2) is facilitated by Tamm-Horsfall protein in a chloride-sensitive manner. J Biol Chem 2011;286:30200-10. [Crossref] [PubMed]
- Graham LA, Padmanabhan S, Fraser NJ, et al. Validation of uromodulin as a candidate gene for human essential hypertension. Hypertension 2014;63:551-8. [Crossref] [PubMed]
- Trudu M, Janas S, Lanzani C, et al. Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat Med 2013;19:1655-60. [Crossref] [PubMed]
- Ren M, Ng FL, Warren HR, et al. The biological impact of blood pressure-associated genetic variants in the natriuretic peptide receptor C gene on human vascular smooth muscle. Hum Mol Genet 2018;27:199-210. [Crossref] [PubMed]
- Ng FL, Boedtkjer E, Witkowska K, et al. Increased NBCn1 expression, Na+/HCO3- co-transport and intracellular pH in human vascular smooth muscle cells with a risk allele for hypertension. Hum Mol Genet 2017;26:989-1002. [PubMed]
- Zhang R, Witkowska K, Afonso Guerra-Assuncao J, et al. A blood pressure-associated variant of the SLC39A8 gene influences cellular cadmium accumulation and toxicity. Hum Mol Genet 2016;25:4117-26. [Crossref] [PubMed]
- Nakao E, Adachi H, Enomoto M, et al. Elevated Plasma Transforming Growth Factor beta1 Levels Predict the Development of Hypertension in Normotensives: The 14-Year Follow-Up Study. Am J Hypertens 2017;30:808-14. [Crossref] [PubMed]
- Truty MJ, Lomberk G, Fernandez-Zapico ME, et al. Silencing of the transforming growth factor-beta (TGFbeta) receptor II by Kruppel-like factor 14 underscores the importance of a negative feedback mechanism in TGFbeta signaling. J Biol Chem 2009;284:6291-300. [Crossref] [PubMed]
- Rakyan VK, Down TA, Balding DJ, et al. Epigenome-wide association studies for common human diseases. Nat Rev Genet 2011;12:529-41. [Crossref] [PubMed]
- Richard MA, Huan T, Ligthart S, et al. DNA Methylation Analysis Identifies Loci for Blood Pressure Regulation. Am J Hum Genet 2017;101:888-902. [Crossref] [PubMed]
- Zhao W, Rasheed A, Tikkanen E, et al. Identification of new susceptibility loci for type 2 diabetes and shared etiological pathways with coronary heart disease. Nat Genet 2017;49:1450-7. [Crossref] [PubMed]
- Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet 2012;44:981-90. [Crossref] [PubMed]
- Smith EN, Chen W, Kahonen M, et al. Longitudinal genome-wide association of cardiovascular disease risk factors in the Bogalusa heart study. PLoS Genet 2010;6. [Crossref] [PubMed]
- Nagy R, Boutin TS, Marten J, et al. Exploration of haplotype research consortium imputation for genome-wide association studies in 20,032 Generation Scotland participants. Genome Med 2017;9:23. [Crossref] [PubMed]
- Surakka I, Horikoshi M, Magi R, et al. The impact of low-frequency and rare variants on lipid levels. Nat Genet 2015;47:589-97. [Crossref] [PubMed]
- Pietiainen V, Lindgren CM, Stefansson K, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet 2013;45:1274-83. [Crossref] [PubMed]
- Yin XM, Oltvai ZN, Korsmeyer SJ. BH1 and BH2 domains of Bcl-2 are required for inhibition of apoptosis and heterodimerization with Bax. Nature 1994;369:321-3. [Crossref] [PubMed]
- Cangiano E, Marchesini J, Campo G, et al. ACE inhibition modulates endothelial apoptosis and renewal via endothelial progenitor cells in patients with acute coronary syndromes. Am J Cardiovasc Drugs 2011;11:189-98. [Crossref] [PubMed]
- Radha G, Raghavan SC. BCL2: A promising cancer therapeutic target. Biochim Biophys Acta 2017;1868:309-14. [PubMed]
- Summar ML, Gainer JV, Pretorius M, et al. Relationship between carbamoyl-phosphate synthetase genotype and systemic vascular function. Hypertension 2004;43:186-91. [Crossref] [PubMed]
- Plubell DL, Wilmarth PA, Zhao Y, et al. Extended Multiplexing of Tandem Mass Tags (TMT) Labeling Reveals Age and High Fat Diet Specific Proteome Changes in Mouse Epididymal Adipose Tissue. Mol Cell Proteomics 2017;16:873-90. [Crossref] [PubMed]
- McReynolds JW, Crowley B, Mahoney MJ, et al. Autosomal recessive inheritance of human mitochondrial carbamyl phosphate synthetase deficiency. Am J Hum Genet 1981;33:345-53. [PubMed]
- Jockers R, Delagrange P, Dubocovich ML, et al. Update on melatonin receptors: IUPHAR Review 20. Br J Pharmacol 2016;173:2702-25. [Crossref] [PubMed]
- Comai S, Ochoa-Sanchez R, Gobbi G. Sleep-wake characterization of double MT(1)/MT(2) receptor knockout mice and comparison with MT(1) and MT(2) receptor knockout mice. Behav Brain Res 2013;243:231-8. [Crossref] [PubMed]
- Obayashi K, Saeki K, Tone N, et al. Relationship between melatonin secretion and nighttime blood pressure in elderly individuals with and without antihypertensive treatment: a cross-sectional study of the HEIJO-KYO cohort. Hypertens Res 2014;37:908-13. [Crossref] [PubMed]
- Grossman E, Laudon M, Yalcin R, et al. Melatonin reduces night blood pressure in patients with nocturnal hypertension. Am J Med 2006;119:898-902. [Crossref] [PubMed]
- Bonnefond A, Froguel P. Disentangling the Role of Melatonin and its Receptor MTNR1B in Type 2 Diabetes: Still a Long Way to Go? Curr Diab Rep 2017;17:122. [Crossref] [PubMed]
- Scott RA, Scott LJ, Magi R, et al. An Expanded Genome-Wide Association Study of Type 2 Diabetes in Europeans. Diabetes 2017;66:2888-902. [Crossref] [PubMed]
- Nelson CP, Goel A. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet 2017;49:1385-91. [PubMed]
- Nikpay M, Goel A, Won HH, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 2015;47:1121-30. [Crossref] [PubMed]
- Traylor M, Farrall M, Holliday EG, et al. Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies. Lancet Neurol 2012;11:951-62. [Crossref] [PubMed]
- Kottgen A, Pattaro C, Boger CA, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet 2010;42:376-84. [Crossref] [PubMed]
- Smith NL, Felix JF, Morrison AC, et al. Association of genome-wide variation with the risk of incident heart failure in adults of European and African ancestry: a prospective meta-analysis from the cohorts for heart and aging research in genomic epidemiology (CHARGE) consortium. Circ Cardiovasc Genet 2010;3:256-66. [Crossref] [PubMed]
- Warren HR, Evangelou E. Genome-wide association analysis identifies novel blood pressure loci and offers biological insights into cardiovascular risk. Nat Genet 2017;49:403-15. [Crossref] [PubMed]
- Pattaro C, Teumer A. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun 2016;7:10023. [Crossref] [PubMed]
- Manni M, Ricker E, Pernis AB. Regulation of systemic autoimmunity and CD11c(+) Tbet(+) B cells by SWEF proteins. Cell Immunol 2017;321:46-51. [Crossref] [PubMed]
- Sun C, Molineros JE, Looger LL, et al. High-density genotyping of immune-related loci identifies new SLE risk variants in individuals with Asian ancestry. Nat Genet 2016;48:323-30. [Crossref] [PubMed]
- Hippisley-Cox J, Coupland C, Brindle P. Development and validation of QRISK3 risk prediction algorithms to estimate future risk of cardiovascular disease: prospective cohort study. BMJ 2017;357:j2099. [Crossref] [PubMed]
- Dwyer J, Azzi S, Leclair HM, et al. The guanine exchange factor SWAP70 mediates vGPCR-induced endothelial plasticity. Cell Commun Signal 2015;13:11. [PubMed]
- Izzedine H, Ederhy S, Goldwasser F, et al. Management of hypertension in angiogenesis inhibitor-treated patients. Ann Oncol 2009;20:807-15. [Crossref] [PubMed]
- Faruque LI, Lin M, Battistella M, et al. Systematic review of the risk of adverse outcomes associated with vascular endothelial growth factor inhibitors for the treatment of cancer. PLoS One 2014;9. [Crossref] [PubMed]
- Wheeler E, Leong A, Liu CT. Impact of common genetic determinants of Hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: A transethnic genome-wide meta-analysis. PLoS Med 2017;14. [Crossref] [PubMed]
- Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010;466:707-13. [Crossref] [PubMed]
- Reilly MP, Li M, He J, et al. Identification of ADAMTS7 as a novel locus for coronary atherosclerosis and association of ABO with myocardial infarction in the presence of coronary atherosclerosis: two genome-wide association studies. Lancet 2011;377:383-92. [Crossref] [PubMed]
- Whincup PH, Cook DG, Phillips AN, et al. ABO blood group and ischaemic heart disease in British men. BMJ 1990;300:1679-82. [Crossref] [PubMed]
- Chen Z, Yang SH, Xu H, et al. ABO blood group system and the coronary artery disease: an updated systematic review and meta-analysis. Sci Rep 2016;6:23250. [PubMed]
- Gallinaro L, Cattini MG, Sztukowska M, et al. A shorter von Willebrand factor survival in O blood group subjects explains how ABO determinants influence plasma von Willebrand factor. Blood 2008;111:3540-5. [Crossref] [PubMed]
- Jenkins PV, O'Donnell JS. ABO blood group determines plasma von Willebrand factor levels: a biologic function after all? Transfusion 2006;46:1836-44. [Crossref] [PubMed]
- YUGCC. Pleiotropic effect of common variants at ABO Glycosyltranferase locus in 9q32 on plasma levels of pancreatic lipase and angiotensin converting enzyme. PLoS One 2014;9. [Crossref] [PubMed]
- Chung CM, Wang RY, Chen JW, et al. A genome-wide association study identifies new loci for ACE activity: potential implications for response to ACE inhibitor. Pharmacogenomics J 2010;10:537-44. [Crossref] [PubMed]
- Gasso P, Ritter MA, Mas S, et al. Influence of ABO genotype and phenotype on angiotensin-converting enzyme plasma activity. J Renin Angiotensin Aldosterone Syst 2014;15:580-4. [Crossref] [PubMed]
- D'Agostino RB Sr, Vasan RS, Pencina MJ, et al. General cardiovascular risk profile for use in primary care: the Framingham Heart Study. Circulation 2008;117:743-53. [Crossref] [PubMed]
- Hippisley-Cox J, Coupland C, Vinogradova Y, et al. Derivation and validation of QRISK, a new cardiovascular disease risk score for the United Kingdom: prospective open cohort study. BMJ 2007;335:136. [Crossref] [PubMed]
- Lu X, Huang J, Wang L, et al. Genetic predisposition to higher blood pressure increases risk of incident hypertension and cardiovascular diseases in Chinese. Hypertension 2015;66:786-92. [Crossref] [PubMed]
- Pazoki R, Dehghan A, Evangelou E, et al. Genetic Predisposition to High Blood Pressure and Lifestyle Factors: Associations With Midlife Blood Pressure Levels and Cardiovascular Events. Circulation 2018;137:653-61. [Crossref] [PubMed]
- Nierenberg JL, Li C, He J, et al. Blood Pressure Genetic Risk Score Predicts Blood Pressure Responses to Dietary Sodium and Potassium: The GenSalt Study (Genetic Epidemiology Network of Salt Sensitivity). Hypertension 2017;70:1106-12. [Crossref] [PubMed]
- Ganna A, Magnusson PK, Pedersen NL, et al. Multilocus genetic risk scores for coronary heart disease prediction. Arterioscler Thromb Vasc Biol 2013;33:2267-72. [Crossref] [PubMed]
- Iribarren C, Lu M, Jorgenson E, et al. Clinical Utility of Multimarker Genetic Risk Scores for Prediction of Incident Coronary Heart Disease: A Cohort Study Among Over 51 Thousand Individuals of European Ancestry. Circ Cardiovasc Genet 2016;9:531-40. [Crossref] [PubMed]
- Mega JL, Stitziel NO, Smith JG, et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet 2015;385:2264-71. [Crossref] [PubMed]
- Knowles JW, Zarafshar S, Pavlovic A, et al. Impact of a Genetic Risk Score for Coronary Artery Disease on Reducing Cardiovascular Risk: A Pilot Randomized Controlled Study. Front Cardiovasc Med 2017;4:53. [Crossref] [PubMed]
- Kullo IJ, Jouni H, Austin EE, et al. Incorporating a Genetic Risk Score Into Coronary Heart Disease Risk Estimates: Effect on Low-Density Lipoprotein Cholesterol Levels (the MI-GENES Clinical Trial). Circulation 2016;133:1181-8. [Crossref] [PubMed]
- Abegaz TM, Shehab A, Gebreyohannes EA, et al. Nonadherence to antihypertensive drugs: A systematic review and meta-analysis. Medicine (Baltimore) 2017;96. [Crossref] [PubMed]
- Joly Y, Burton H, Knoppers BM, et al. Life insurance: genomic stratification and risk classification. Eur J Hum Genet 2014;22:575-9. [Crossref] [PubMed]
- Li C, He J, Chen J, et al. Genome-Wide Gene-Sodium Interaction Analyses on Blood Pressure: The Genetic Epidemiology Network of Salt-Sensitivity Study. Hypertension 2016;68:348-55. [Crossref] [PubMed]
- Sansanwal P, Kambham N, Sarwal MM. Caspase-4 may play a role in loss of proximal tubules and renal injury in nephropathic cystinosis. Pediatr Nephrol 2010;25:105-9. [Crossref] [PubMed]
- Waskiewicz AJ, Flynn A, Proud CG, et al. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J 1997;16:1909-20. [Crossref] [PubMed]
- Lu D, Yang H, Raizada MK. Angiotensin II regulation of neuromodulation: downstream signaling mechanism from activation of mitogen-activated protein kinase. J Cell Biol 1996;135:1609-17. [Crossref] [PubMed]
- Sung YJ, Winkler TW, de Las Fuentes L, et al. A Large-Scale Multi-ancestry Genome-wide Study Accounting for Smoking Behavior Identifies Multiple Significant Loci for Blood Pressure. Am J Hum Genet 2018;102:375-400. [Crossref] [PubMed]
- Simino J, Sung YJ, Kume R, et al. Gene-alcohol interactions identify several novel blood pressure loci including a promising locus near SLC16A9. Front Genet 2013;4:277. [Crossref] [PubMed]
- Kim YK, Kim Y, Hwang MY, et al. Identification of a genetic variant at 2q12.1 associated with blood pressure in East Asians by genome-wide scan including gene-environment interactions. BMC Med Genet 2014;15:65. [Crossref] [PubMed]
- Falaschetti E, Mindell J, Knott C, et al. Hypertension management in England: a serial cross-sectional study from 1994 to 2011. Lancet 2014;383:1912-9. [Crossref] [PubMed]
- Turner ST, Schwartz GL, Chapman AB, et al. Antihypertensive pharmacogenetics: getting the right drug into the right patient. J Hypertens 2001;19:1-11. [Crossref] [PubMed]
- Materson BJ, Reda DJ, Cushman WC, et al. Single-drug therapy for hypertension in men. A comparison of six antihypertensive agents with placebo. The Department of Veterans Affairs Cooperative Study Group on Antihypertensive Agents. N Engl J Med 1993;328:914-21. [Crossref] [PubMed]
- Wu J, Kraja AT, Oberman A, et al. A summary of the effects of antihypertensive medications on measured blood pressure. Am J Hypertens 2005;18:935-42. [Crossref] [PubMed]
- NICE. Hypertension in adults: diagnosis and management Clinical guideline [CG127]. Available online: https://www.nice.org.uk/guidance/cg127
- Freis ED, Materson BJ, Flamenbaum V. Comparison of propranolol or hydrochlorothiazide alone for treatment of hypertension. III. Evaluation of the renin-angiotensin system. Am J Med 1983;74:1029-41. [Crossref] [PubMed]
- Liu J, Liu ZQ, Yu BN, et al. beta1-Adrenergic receptor polymorphisms influence the response to metoprolol monotherapy in patients with essential hypertension. Clin Pharmacol Ther 2006;80:23-32. [Crossref] [PubMed]
- Muthumala A, Drenos F, Elliott PM, et al. Role of beta adrenergic receptor polymorphisms in heart failure: systematic review and meta-analysis. Eur J Heart Fail 2008;10:3-13. [Crossref] [PubMed]
- Gong Y, McDonough CW, Wang Z, et al. Hypertension susceptibility loci and blood pressure response to antihypertensives: results from the pharmacogenomic evaluation of antihypertensive responses study. Circ Cardiovasc Genet 2012;5:686-91. [Crossref] [PubMed]
- Svensson-Farbom P, Wahlstrand B, Almgren P, et al. A functional variant of the NEDD4L gene is associated with beneficial treatment response with beta-blockers and diuretics in hypertensive patients. J Hypertens 2011;29:388-95. [Crossref] [PubMed]
- Turner ST, Bailey KR, Fridley BL, et al. Genomic association analysis suggests chromosome 12 locus influencing antihypertensive response to thiazide diuretic. Hypertension 2008;52:359-65. [Crossref] [PubMed]
- Turner ST, Boerwinkle E, O'Connell JR, et al. Genomic association analysis of common variants influencing antihypertensive response to hydrochlorothiazide. Hypertension 2013;62:391-7. [Crossref] [PubMed]
- Gong Y, Wang Z, Beitelshees AL, et al. Pharmacogenomic Genome-Wide Meta-Analysis of Blood Pressure Response to beta-Blockers in Hypertensive African Americans. Hypertension 2016;67:556-63. [PubMed]
- Turner ST, Bailey KR, Schwartz GL, et al. Genomic association analysis identifies multiple loci influencing antihypertensive response to an angiotensin II receptor blocker. Hypertension 2012;59:1204-11. [Crossref] [PubMed]
- Hiltunen TP, Donner KM, Sarin AP, et al. Pharmacogenomics of hypertension: a genome-wide, placebo-controlled cross-over study, using four classes of antihypertensive drugs. J Am Heart Assoc 2015;4. [Crossref] [PubMed]
- Mas S, Gasso P, Alvarez S, et al. Pharmacogenetic predictors of angiotensin-converting enzyme inhibitor-induced cough: the role of ACE, ABO, and BDKRB2 genes. Pharmacogenet Genomics 2011;21:531-8. [Crossref] [PubMed]
- Luo JQ, He FZ, Luo ZY, et al. Rs495828 polymorphism of the ABO gene is a predictor of enalapril-induced cough in Chinese patients with essential hypertension. Pharmacogenet Genomics 2014;24:306-13. [Crossref] [PubMed]
- Luo JQ, He FZ, Wang ZM, et al. SLCO1B1 Variants and Angiotensin Converting Enzyme Inhibitor (Enalapril)-Induced Cough: a Pharmacogenetic Study. Sci Rep 2015;5:17253. [Crossref] [PubMed]
- Mosley JD, Shaffer CM, Van Driest SL, et al. A genome-wide association study identifies variants in KCNIP4 associated with ACE inhibitor-induced cough. Pharmacogenomics J 2016;16:231-7. [Crossref] [PubMed]
- Kim TB, Oh SY, Park HK, et al. Polymorphisms in the neurokinin-2 receptor gene are associated with angiotensin-converting enzyme inhibitor-induced cough. J Clin Pharm Ther 2009;34:457-64. [Crossref] [PubMed]
- Li YF, Zhu XM, Liu F, et al. Angiotensin-converting enzyme (ACE) gene insertion/deletion polymorphism and ACE inhibitor-related cough: a meta-analysis. PLoS One 2012;7. [Crossref] [PubMed]
- Hallberg P, Persson M, Axelsson T, et al. Genetic variants associated with angiotensin-converting enzyme inhibitor-induced cough: a genome-wide association study in a Swedish population. Pharmacogenomics 2017;18:201-13. [Crossref] [PubMed]
- Ware JS, Wain LV, Channavajjhala SK, et al. Phenotypic and pharmacogenetic evaluation of patients with thiazide-induced hyponatremia. J Clin Invest 2017;127:3367-74. [Crossref] [PubMed]
- Vandell AG, McDonough CW, Gong Y, et al. Hydrochlorothiazide-induced hyperuricaemia in the pharmacogenomic evaluation of antihypertensive responses study. J Intern Med 2014;276:486-97. [Crossref] [PubMed]
- Mallal S, Phillips E, Carosi G, et al. HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med 2008;358:568-79. [Crossref] [PubMed]
- Arnett DK, Boerwinkle E, Davis BR, et al. Pharmacogenetic approaches to hypertension therapy: design and rationale for the Genetics of Hypertension Associated Treatment (GenHAT) study. Pharmacogenomics J 2002;2:309-17. [Crossref] [PubMed]
- Salvi E, Wang Z, Rizzi F, et al. Genome-Wide and Gene-Based Meta-Analyses Identify Novel Loci Influencing Blood Pressure Response to Hydrochlorothiazide. Hypertension 2017;69:51-9. [Crossref] [PubMed]
- Bis JC, Sitlani C, Irvin R, et al. Drug-Gene Interactions of Antihypertensive Medications and Risk of Incident Cardiovascular Disease: A Pharmacogenomics Study from the CHARGE Consortium. PLoS One 2015;10. [Crossref] [PubMed]
- Roden DM. Phenome-wide association studies: a new method for functional genomics in humans. J Physiol 2017;595:4109-15. [Crossref] [PubMed]


