The impact of GAB2 genetic variations on cerebrospinal fluid markers in Alzheimer’s disease
Introduction
Alzheimer’s disease (AD) can explain about 59% of all dementia, which is deemed as the leading form of dementia among elderly (1). It is believed that genetic and environmental factors influence the incidence of the late-onset AD (LOAD) together. Growth factor receptor-bound protein-associated binding protein 2 (GAB2), as well as other susceptibility genes, had been confirmed to have significant associations with the LOAD by several genome wide association studies (GWAS) (2-6). GAB2, situated on 11q14.1, was first discovered to modify AD risk in APOE ε4 carriers in Caucasian (7). Since then, subsequent research was continually conducted to explore the association between GAB2 gene and the AD susceptibility in various districts and ethnic groups, and controversial conclusions were obtained (8-21).
GAB2 protein belongs to the family of scaffolding and adapter proteins, and is abundantly expressed in the central nervous system (CNS) especially in prefrontal cortex and hypothalamus. It participates in many signaling pathways including cell proliferation, differentiation, survival and apoptosis (22,23). In AD, GAB2 protein is an amplifier of PI3K/AKT signaling, which is able to reduce tau phosphorylation to prevent the formation of neurofibrillary tangles (NFTs) and neuronal loss (24,25). The diminished GAB2 expression would lead to a high level of phosphorylation of tau and the formation of NFTs, which are induced by glycogen synthase kinase 3β (Gsk-3β). Reiman et al. also demonstrated small interfering RNA (siRNA)-mediated reduction of GAB2 protein increased tau phosphorylation and NFTs formation in vitro (7). From the evidence, GAB2 can alter tau pathology and further mediate the susceptibility of AD. However, it is still undefined about the role of GAB2 polymorphisms. As phosphorylated tau (P-tau), total tau (T-tau) and amyloid β (Aβ) in cerebrospinal fluid (CSF) could be effective biomarkers of AD; however, little is illustrated whether GAB2 polymorphisms mediated the susceptibility of AD through the change of CSF biomarkers. Therefore, in our study, we research the impact of GAB2 genetic polymorphisms on CSF biomarkers. In this way, we sought to provide clues to how GAB2 genetic polymorphisms influence AD pathogenesis.
Methods
Alzheimer’s Disease Neuroimaging Initiative (ADNI) dataset and subjects
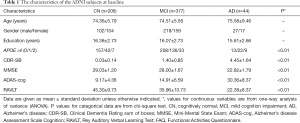
In our paper, we used the data from the ADNI database (https://ida.loni.usc.edu/), which is a large, multicenter, longitudinal neuroimaging study. At the very beginning, ADNI aimed to identify mild cognitive impairment (MCI) and AD at earlier stages under the combination of serial magnetic resonance imaging (MRI), other biological markers, and clinical and neuropsychological assessment. The details about the ADNI cohort were described elsewhere (26,27). Our study recruited 627 individuals, including 206 cognitively normal (CN), 377 MCI patients, and 44 AD patients at baseline eventually and we downloaded their detailed demographics from the ADNI website in 2015 (Table 1). The ADNI study was approved by all the Institutional Ethical Review Boards of all participating centers. All participants signed written informed consent.
Full table
Single nucleotide polymorphisms (SNPs) selection
We extracted GAB2 genotypes from the ADNI PLINK format data and performed the quality control (QC) procedures with the use of PLINK v1.07 (28).The following were the inclusion criteria: minimum call rates >90%, minimum minor allele frequencies (MAF) >0.01, Hardy-Weinberg equilibrium test P>1×10−3. Finally, with the method of Haploview 4.2 platform, four targeted GAB2 loci (rs1385600, rs1007837, rs10793294, rs2373115) were selected in our research.
CSF proteins
We downloaded all CSF data from ADNI dataset. The steps of acquisition and measurement of CSF were as follow: firstly, collected and then transferred samples into corresponding tubes; secondly, held them in dry ice for 1 hour for freezing; thirdly, transported them to corresponding laboratory at the University of Pennsylvania Medical Center for measurement; fourthly, thawed them at room temperature, prepared aliquots (0.5 mL)and reserved them at −80 °C; finally, measured proteins with the multiplex xMAP Luminex platform and with Innogenetics immunoassay kit-based reagents (29). Full details of analytical platform are exhibited at site (http://adni.loni.ucla.edu).
Statistical analysis
We carried out all statistical analyses with R3.12 (http://www.r-project.org/) and PLINK 1.07 (http://pngu.mgh.harvard.edu/wpurcell/plink/). One-way analysis of variance (ANOVA) was used to test differences in continuous variables (age, education years, cognitive scores, volume, etc.) and chi-square test was used to examine categorical data (gender, APOE ε4 status). We assessed possible relationship between GAB2 SNPs and CSF biomarkers at baseline in total sample by a multiple linear regression model, in which age, gender, education years, and APOE ε4 status were regarded as covariates. In addition, we explored the correlation between these GAB2 SNPs and these suggestive phenotypes in the haplotype-based association analysis, and made subgroup analysis to investigate the influence of GAB2 loci in the AD pathogenesis. We used Bonferroni correction to control the false discovery rate of multiple tests. Bonferroni-corrected Pc<0.05 was considered statistically significant.
Results
Characteristics of included subjects
We enrolled 206 CN (104 women, 74.36±5.79 years), 377 MCI (159 women, 74.51±5.56 years) and 44 AD patients (17 women, 75.58±9.46 years) in this study and Table 1 presented the characteristics of the included subjects. Age, gender and education were matched between the three subgroups. As would be expected, the frequency of the APOE ε4 allele is highest in AD group (45.5%) compared to the MCI (26.8%) and CN (13.6%) subjects. The cognitive function of all subjects was measured by various neuropsychological scales (Table 1). Unsurprisingly, the worst performance was displayed by AD patients.
CSF markers and GAB2 genotypes
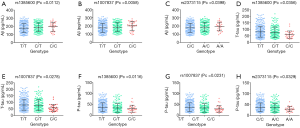
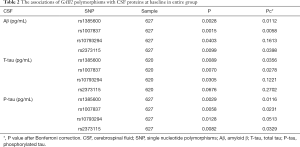
We firstly confirmed relations between CSF biomarker sand GAB2 genotypes. There were significant relations between all the three CSF biomarkers and GAB2 loci (Figure 1, Table 2). We discovered that both rs1385600 and rs1007837 were significantly associated with all the three biomarkers in CSF (rs1385600: Aβ Pc =0.0112, T-tau Pc =0.0356, P-tau Pc =0.0116; rs1007837: Aβ Pc =0.0058, T-tau Pc =0.0278, P-tau Pc =0.0231), whereas rs2373115 only showed significant association with Aβ and P-tau (Aβ Pc =0.0398, P-tau Pc =0.0329). However, the levels of all the three biomarkers are not significantly different among the three genotypes of rs10793294.
Full table
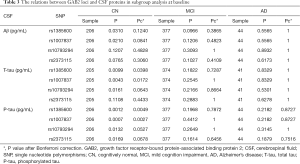
Additionally, subgroup analysis was performed to make sure whether GAB2 genetic polymorphisms regulated the levels of CSF markers in AD, MCI and CN subgroup (Table 3). After Bonferroni correction, only rs1385600 and rs1007837 showed significant associations with the expression of P-tau and T-tau in CN subgroup (rs1385600: T-tau Pc =0.0398, P-tau Pc =0.0049; rs1007837: T-tau Pc =0.0172, P-tau Pc =0.0027). However, none of these loci changed T-tau and P-tau expression in MCI and AD subgroup. Likewise, we did not observe any GAB2 loci which altered the levels of Aβ in AD, MCI and CN subgroup (Table 3).
Full table
Discussion
Our study has analyzed the correlations between GAB2 genotypes and CSF biomarkers in ADNI database. We corroborated that GAB2 genetic variants displayed significant effects in the expression of the three CSF biomarkers.
Different with many prior findings about the involvement of GAB2 in AD pathogenesis, our study revealed that GAB2 loci altered the expression of Aβ, P-tau and T-tau. Among the four SNPs associated with the level of CSF proteins, rs2373115 has also been showed a significant association with AD in APOE ε4 carriers (30). Rs2373115 is the locus that has been studied most frequently in different races followed by rs1385600 and rs1007837, and various conclusions were drawn. Interestingly, Zou et al. considered rs1385600 and rs2373115 as protective GAB2 variants because they could reduce LOAD pathology via the elevated GAB2 mRNA levels in lymphoblastoid cells in some ethnicities. Additionally, we should pay attention to that we did not confirm any association of the four SNPs with the three CSF biomarkers in MCI and AD subgroup. This might be explained by the different roles of these loci in CN individual verses those in MCI and AD.
GAB2 protein plays a vital role in the phosphorylation of kinases that involve in core neuropathogenesis of AD (7,31-33). The abnormal tau is thought as one of primary pathological factors in AD. GAB2 protein is responsible for the inhibition of tau protein phosphorylation and tangle formation known as NFTs (7). GAB2 can activate PI3K, resulting in the activation of PKB and further inactivation of GSK-3β (34). GSK-3β overexpression is involved in the phosphorylation of tau, the linkage between amyloid and tau pathology, and microglia-mediated inflammation (35,36). Therefore, it is rational to suppose that the reduced GAB2 expression and/or function would lead to the increase in tau phosphorylation. Moreover, studies in vitro indicated that the diminished GAB2 protein working through siRNA could increase tau phosphorylation (7). It remains unclear whether this coupling affects phosphor-ERK-1 and phosphor-ERK-2 signal transduction pathway though this mechanism in AD pathogenesis (37). In addition, GAB2 also promotes neural/glial cell proliferation to prevent the occurrence of AD via the interaction with a Rho family GTPase-activating protein (GC-GAP) which is mainly expressed in the brain (38). Collectively, GAB2 is a scaffolding protein linked with multiple signaling pathways that influence AD-related tau and cell survival or other aspects of AD pathogenesis.
Now, the genetic effects on neuroimaging and CSF markers can be depicted directly using neuroimaging phenotypes (39). So, possible mechanisms through which these genetic factors modulate the development of AD might be disclosed. The crucial strength of this study lies in quantitative traits (QTs) association studies, which have less sample size and increased statistical power requirements compared to traditional case-control designs. Moreover, ADNI database has advantages of detailed cognitive assessment agreement and diagnostic criteria, and shows relative changes across the spectrum of AD. However, we should notice that there still are some limitations. Firstly, the QT analysis reduced the sample size, because not all the participants have all information. Secondly, we recruited only Caucasians to reduce the influence of population stratification. However, the frequency of four GAB2 loci differs in various populations, so our results were not on behalf of other ethnicities and replication studies in other races are indispensable. Thirdly, not all candidate gene variants in GAB2 of ADNI database were analyzed.
Collectively, our research demonstrated that GAB2 variants were significantly associated with the expression of Aβ, T-tau and P-tau protein in CSF, which further supported that GAB2 genetic variation modulates AD risk via the alteration of both Aβ and tau pathology. Moreover, these variants might specifically affect CN individuals, which offer new insights into the relationship between GAB2 polymorphisms and AD. However, several limitations precluded the explanation of these findings, and future researches are wanted to figure out the detailed molecular mechanisms by which GAB2 influence Aβ and tau protein with a longer follow-up or in other ethnicities, and an independent population is needed to replicate these findings.
Acknowledgements
The authors thank scientists contributed in developing the clinical and genetic resources necessary to collect these data and complete this project. The authors also gratefully thank the efforts of hundreds of individuals whose help and participation made this work possible.
Funding: Data collection and sharing for this project was funded by the AD Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The ADNI study was approved by all the Institutional Ethical Review Boards of all participating centers. All participants signed written informed consent.
References
- Holtzman DM, Morris JC, Goate AM. Alzheimer's disease: the challenge of the second century. Sci Transl Med 2011;3:77sr1. [Crossref] [PubMed]
- Naj AC, Jun G, Beecham GW, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer's disease. Nat Genet 2011;43:436-41. [Crossref] [PubMed]
- Hollingworth P, Harold D, Sims R, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet 2011;43:429-35. [Crossref] [PubMed]
- Seshadri S, Fitzpatrick AL, Ikram MA, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 2010;303:1832-40. [Crossref] [PubMed]
- Lambert JC, Heath S, Even G, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet 2009;41:1094-9. [Crossref] [PubMed]
- Harold D, Abraham R, Hollingworth P, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet 2009;41:1088-93. [Crossref] [PubMed]
- Reiman EM, Webster JA, Myers AJ, et al. GAB2 alleles modify Alzheimer's risk in APOE epsilon4 carriers. Neuron 2007;54:713-20. [Crossref] [PubMed]
- Jiao B, Liu X, Zhou L, et al. Polygenic Analysis of Late-Onset Alzheimer's Disease from Mainland China. PLoS One 2015;10:e0144898. [Crossref] [PubMed]
- Zhong XL, Yu JT, Hou GY, et al. Common variant in GAB2 is associated with late-onset Alzheimer's disease in Han Chinese. Clin Chim Acta 2011;412:446-9. [Crossref] [PubMed]
- Wang G, Pan XL, Cui PJ, et al. Association study of the GAB2 gene with the risk of Alzheimer disease in the chinese population. Alzheimer Dis Assoc Disord 2011;25:283-5. [Crossref] [PubMed]
- Zou F, Belbin O, Carrasquillo MM, et al. Linking protective GAB2 variants, increased cortical GAB2 expression and decreased Alzheimer's disease pathology. PLoS One 2013;8:e64802. [Crossref] [PubMed]
- Jin C, Wu CZ, Liu X, et al. GAB2 polymorphism rs2373115 confers susceptibility to sporadic Alzheimer's disease. Neurosci Lett 2013;556:216-20. [Crossref] [PubMed]
- Izzo G, Forlenza OV, Santos B, et al. Single-nucleotide polymorphisms of GSK3B, GAB2 and SORL1 in late-onset Alzheimer's disease: interactions with the APOE genotype. Clinics (Sao Paulo) 2013;68:277-80. [Crossref] [PubMed]
- Belbin O, Carrasquillo MM, Crump M, et al. Investigation of 15 of the top candidate genes for late-onset Alzheimer's disease. Hum Genet 2011;129:273-82. [Crossref] [PubMed]
- Zhou S. Association analysis between polymorphism rs2373115 of gene GRB-associated binding protein 2 and Mongolian Alzheimer patients. Neurosci Lett 2010;480:4-6. [Crossref] [PubMed]
- Lin K, Tang M, Han H, et al. GAB2 is not associated with late-onset Alzheimer's disease in Chinese Han. Neurol Sci 2010;31:277-81. [Crossref] [PubMed]
- Laumet G, Chouraki V, Grenier-Boley B, et al. Systematic analysis of candidate genes for Alzheimer's disease in a French, genome-wide association study. J Alzheimers Dis 2010;20:1181-8. [Crossref] [PubMed]
- Ramirez-Lorca R, Boada M, Saez ME, et al. GAB2 gene does not modify the risk of Alzheimer's disease in Spanish APOE 4 carriers. J Nutr Health Aging 2009;13:214-9. [Crossref] [PubMed]
- Nacmias B, Tedde A, Bagnoli S, et al. Implication of GAB2 gene polymorphism in Italian patients with Alzheimer's disease. J Alzheimers Dis 2009;16:513-5. [Crossref] [PubMed]
- Miyashita A, Arai H, Asada T, et al. GAB2 is not associated with late-onset Alzheimer's disease in Japanese. Eur J Hum Genet 2009;17:682-6. [Crossref] [PubMed]
- Chapuis J, Hannequin D, Pasquier F, et al. Association study of the GAB2 gene with the risk of developing Alzheimer's disease. Neurobiol Dis 2008;30:103-6. [Crossref] [PubMed]
- Sarmay G, Angyal A, Kertesz A, et al. The multiple function of Grb2 associated binder (Gab) adaptor/scaffolding protein in immune cell signaling. Immunol Lett 2006;104:76-82. [Crossref] [PubMed]
- Liu Y, Rohrschneider LR. The gift of Gab. FEBS Lett 2002;515:1-7. [Crossref] [PubMed]
- Hibar DP, Jahanshad N, Stein JL, et al. Alzheimer's disease risk gene, GAB2, is associated with regional brain volume differences in 755 young healthy twins. Twin Res Hum Genet 2012;15:286-95. [Crossref] [PubMed]
- Sugden PH, Fuller SJ, Weiss SC, et al. Glycogen synthase kinase 3 (GSK3) in the heart: a point of integration in hypertrophic signalling and a therapeutic target? A critical analysis. Br J Pharmacol 2008;153 Suppl 1:S137-53. [Crossref] [PubMed]
- Petersen RC, Aisen PS, Beckett LA, et al. Alzheimer's Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology 2010;74:201-9. [Crossref] [PubMed]
- Mueller SG, Weiner MW, Thal LJ, et al. The Alzheimer's disease neuroimaging initiative. Neuroimaging Clin N Am 2005;15:869-77. xi-xii. [Crossref] [PubMed]
- Saykin AJ, Shen L, Foroud TM, et al. Alzheimer's Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimers Dement 2010;6:265-73. [Crossref] [PubMed]
- Olsson A, Vanderstichele H, Andreasen N, et al. Simultaneous measurement of beta-amyloid(1-42), total tau, and phosphorylated tau (Thr181) in cerebrospinal fluid by the xMAP technology. Clin Chem 2005;51:336-45. [Crossref] [PubMed]
- Ikram MA, Liu F, Oostra BA, et al. The GAB2 gene and the risk of Alzheimer's disease: replication and meta-analysis. Biol Psychiatry 2009;65:995-9. [Crossref] [PubMed]
- Sleegers K, Bettens K, Brouwers N, et al. Common variation in GRB-associated Binding Protein 2 (GAB2) and increased risk for Alzheimer dementia. Hum Mutat 2009;30:E338-44. [Crossref] [PubMed]
- Nizzari M, Venezia V, Repetto E, et al. Amyloid precursor protein and Presenilin1 interact with the adaptor GRB2 and modulate ERK 1,2 signaling. J Biol Chem 2007;282:13833-44. [Crossref] [PubMed]
- Russo C, Dolcini V, Salis S, et al. Signal transduction through tyrosine-phosphorylated carboxy-terminal fragments of APP via an enhanced interaction with Shc/Grb2 adaptor proteins in reactive astrocytes of Alzheimer's disease brain. Ann N Y Acad Sci 2002;973:323-33. [Crossref] [PubMed]
- Baki L, Shioi J, Wen P, et al. PS1 activates PI3K thus inhibiting GSK-3 activity and tau overphosphorylation: effects of FAD mutations. EMBO J 2004;23:2586-96. [Crossref] [PubMed]
- Hernandez F, Gomez de Barreda E, Fuster-Matanzo A, et al. GSK3: a possible link between beta amyloid peptide and tau protein. Exp Neurol 2010;223:322-5. [Crossref] [PubMed]
- Hernandez F, de Barreda EG, Fuster-Matanzo A, et al. The role of GSK3 in Alzheimer disease. Brain Res Bull 2009;80:248-50. [Crossref] [PubMed]
- Venezia V, Nizzari M, Repetto E, et al. Amyloid precursor protein modulates ERK-1 and -2 signaling. Ann N Y Acad Sci 2006;1090:455-65. [Crossref] [PubMed]
- Zhao C, Ma H, Bossy-Wetzel E, et al. GC-GAP, a Rho family GTPase-activating protein that interacts with signaling adapters Gab1 and Gab2. J Biol Chem 2003;278:34641-53. [Crossref] [PubMed]
- Jagust WJ, Landau SM. Apolipoprotein E, not fibrillar beta-amyloid, reduces cerebral glucose metabolism in normal aging. J Neurosci 2012;32:18227-33. [Crossref] [PubMed]


