Emerging application of genomics-guided therapeutics in personalized lung cancer treatment
Introduction
Despite tremendous effort to fight lung cancer, it remains the leading cause of global cancer-related mortality (1). However, recent advancement in cancer genomics has fueled a revolutionary improvement in our understanding of the driver molecular alterations responsible for tumor progression. The advent of highly sensitive and accurate technological platforms, coupled with a growing body of knowledge, accrued via coordinated global efforts, has opened up new opportunities for designing better therapeutics. Increasingly the focus for treating patients is shifting from conventional cytotoxic chemotherapy to more personalized and targeted precision medicine treatments. Hence, targeted inactivation of driver oncogenes and selective enhancement of host tumor surveillance response are more widespread mode of treatments. For example, in non-small cell lung cancers (NSCLC) activating alterations in the epidermal growth factor receptor (EGFR) or serine/threonine-protein kinase b-raf (BRAF) and chromosomal rearrangements in the anaplastic lymphoma kinase (ALK) or ROS proto-oncogene receptor tyrosine kinase (ROS1) are now clinically-validated targets for kinase-inhibitor therapy (2). Moreover, in recent years, accumulating evidences indicate encouraging future for immunotherapy in treating lung cancer (3,4). Inhibition of CTLA-4, PD-1 and PD-L1 immunosuppressive T cell receptors enhances host immunosurveillance response against tumors for eventual elimination. These successes have ushered in promises for additional targeted therapies in other oncogenic driver subtypes of lung cancer many of which are currently under investigation (5).
Despite remarkable early remission and overall improvement of patient outcome, resistance to targeted therapy invariably occurs. Resistance to targeted therapy can be sub classified into three distinct classes as intrinsic resistance, adaptive resistance and acquired resistance (6). When tumors fail to respond to initial treatment it is defined as intrinsic resistance. For example, tumors harboring EGFR exon 20 deletion failed to respond to EGFR tyrosine kinase inhibitors (TKIs) due to additional preexisting mutations resulting in dysfunction of pro-apoptotic BCL2L11 protein (7). Conversely, resistance against targeted therapy might occur due to a de novo adaptation of cellular epigenetic and transcription programs leading to adaptive resistance and a partial response to the therapy (8). Acquired resistance, on the other hand, arises due to the selective pressure imposed by therapy onto tumor cells consisting of heterogeneous genetic alterations and due to acquisition of therapy induced de novo alterations (6,9). Although, the mechanistic basis of the existence of biological overlap amongst these sub-classes is apparent, detailed mechanistic insights underlying the therapy induced state transitions is currently lacking (6).
This incomplete and short-lived nature of the targeted therapy response gave rise to the idea of a residual disease state in tumor, which is unaffected by the targeted therapy and serves as a prelude for subsequent tumor progression and acquired resistance. A growing body of literature indicates that this evolution of tumors through these different stages is dictated by a heterogeneous combination of multiple molecular drivers (10). Hence, there remains an open question as to how to best study intra and inter-tumor heterogeneity and how to best account for it to strategize improved treatment options. Genomic approaches, in this regard, are guiding cutting-edge lung cancer therapeutics and helping to strategize and predict responses. In this review, we discuss the promises and challenges associated with this genomics-based approach.
Molecular profiling of lung cancer
The advent of new technologies such as massively parallel sequencing and an improvement in our ability to rapidly analyze large datasets have enabled us to profile thousands of tumors and map alterations with genomic resolution, such as mutations, copy number, gene expression, promoter methylation, protein expression and metabolic activity, in tumors (11). Such analyses have led not only to discovery of new biomarkers, to classify tumors into distinct subclasses and therapy regimens, but also to better predict therapy response (12).
Lung cancer, similar to any other cancers, accumulates somatic mutations over time and only a subset of these alterations is considered driver mutations due to their active role in cancer development and progression; the others have been deemed coincidental passengers occurring during the process of tumor evolution (12). Hence, to trace and account for sentinel driver mutations and biomarkers, a concerted effort was put forth by The Cancer Genome Atlas (TCGA) research consortium. They profiled differential somatic mutations between tumor and matched pair normal materials from patients and analyzed tumor DNA, RNA and proteome to identify driver genes and reliable biomarkers for both NSCLC and small cell lung cancer (SCLC). Molecular profiling of 230 resected lung adenocarcinomas from all major histologic types indicated significantly recurrent mutation in a number of 18 genes of which TP53, STK11, KEAP1, NF1, RBM10 and SMARCA4 were of tumor suppressor nature whereas KRAS, EGFR, BRAF and PIK3CA were of oncogenic nature. EGFR was frequently mutated, whereas KRAS was rarely mutated in tumors with high transversion mutation—a mutational signature associated with tobacco smoking. Additionally, the presence of mutual exclusivity between mutant EGFR, KRAS, and BRAF was identified, which reinforced the concept of oncogene driven distinct subclasses of lung cancer. Interestingly, mutual exclusivity was also identified between NF1 tumor suppressor mutation and the KRAS, BRAF and EGFR oncogenes which indicated NF1 loss as a novel driver event for a subclass of lung adenocarcinoma (1). Furthermore, analysis of transcriptome coupled with DNA copy number data identified aberrant copy number alterations of ALK, ROS1 and RET in a small but clinically relevant subset of NSCLC patients (13).
To guide clinical care of lung cancer, molecular genotyping is now a customary practice for lung adenocarcinoma patients. Previous clinical trials have demonstrated efficacy of targeted kinase inhibitors against multiple driver oncogenes, including BRAF and EGFR and against ALK and ROS1 gene fusions (14,15). While many patients benefit initially from this targeted kinase inhibitions, the majorities of responses are incomplete and eventually give rise to drug resistant disease. Predicting the course of resistance can be highly complicated due to intrinsic heterogeneity in the tumor (16). For example, EGFR alterations, found in approximately 15% of U.S. cases, often co-occur with additional activating mutations in CDK4/6, CTNNB1 and PIK3CA (10). Work from Blakely et al. suggests that the presence of these co-occurring mutations dictate not only the aggressiveness of the tumors but also the response of tumors to therapy. Lung adenocarcinomas also often harbor loss-of-function mutations and deletions in tumor suppressor genes such as TP53, STK11, RB1, NF1, CDKN2A, SMARCA4, and KEAP1 (17-19). Unfortunately, such alterations are difficult to exploit therapeutically. Therefore, knowledge of additional genes altered in lung adenocarcinoma is needed to further guide diagnosis and treatment (11).
A similar venture to catalog alterations in SCLC revealed a more complex picture with no singular driver in play. The TCGA analysis from 152 fresh-frozen clinical SCLC tumor specimens demonstrated high genetic heterogeneity, mutational burden and an almost universal bi-allelic inactivation of p53 and RB1 in this subtype (12). Copy number analysis indicated homozygous losses in the CDKN2A locus and amplification of the MYC family genes and tyrosine kinases FGFR1, IRS2 as recurrent genomic events. Additionally, close to half of the tumors examined, had alterations in genes that regulate squamous differentiation e.g., TP63 and SOX2 (20). Further search for relevant SCLC associated mutations, established in previous studies using murine and human models, identified largely mutually exclusive mutations in TP73, RBL1, RBL2 and NOTCH family genes validating their pro-tumorigenic roles on SCLC. Therapeutic agents targeting many of the afore-mentioned candidate genes are being currently pursued clinically (21). Limited available treatment options for SCLC highlights the importance of molecular profiling to identifying novel therapeutic targets.
Molecular profiling has also indicated intrinsic genomic instability in tumors. Tumor mutational burden is also thought to predict response to immunotherapy in NSCLC (22). In theory, increased mutational burden on tumor cells should facilitate the presentation of a greater number of foreign antigens to immune cells. Recent publications suggest, in NSCLC, that the mutational landscape can successfully predict response to immunotherapy (23,24). This opens up an exciting opportunity for targeted immunotherapy in combination with other modes of treatment.
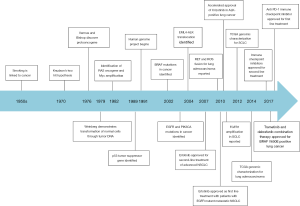
Many successful examples of biomarker driven targeted inhibitors and immunotherapy exemplifies the importance of molecular profiling (Figure 1). In the following section we highlight the evolution of some of the major genomics driven therapeutics.
Precision medicine
We live in the post-genomic era of the cancer therapeutics. Improvements in molecular profiling of individual tumors have prompted a shift away from the use of conventional cytotoxic chemotherapy and towards molecularly targeted agents that include both signal transduction inhibitors (e.g., EGFR, BRAF inhibitors) and immunotherapies (e.g., PD1 and PDL1 antibodies).
Targeted inhibitors in NSCLC
Targeted inhibitors are a remarkable tool for treating NSCLC patients as NSCLC tumors are relatively less sensitive to chemotherapy (25). The most common types of NSCLC are adenocarcinoma, squamous cell carcinoma and large cell carcinoma, of which adenocarcinomas account for approximately 40% of all lung cancers (26). Many examples of targeted intervention in lung adenocarcinoma (LAC) and squamous cell carcinoma (SCLC) exist. Below is a summary of the most notable examples.
Lung adenocarcinoma (LAC)
EGFR
The epidermal growth factor receptor (EGFR; ErbB-1; HER1 in humans) is a transmembrane receptor tyrosine kinase that regulates cellular growth and differentiation in response to EGF family of extracellular protein ligands. EGFR dependent growth signaling is mediated through three distinct downstream signaling arms-MAPK, PI3K/AKT and JAK-STAT pathway.
Somatic alterations in EGFR are present in around 15% of the NSCLC patients and EGFR is one of the major driver oncogenes (1). Currently there are multiple EGFR TKIs approved by FDA. Their response rates are within 50–80%. The first-generation EGFR inhibitors, erlotinib and gefitinib, block EGFR activity in a non-covalent manner, whereas the second and third generation inhibitors, afatinib and osimertinib respectively, block in covalent manner. Osimertinib also spares the wild type EGFR and is emerging as a game-changer for specifically targeting the T790M EGFR variant that causes resistance to first-generation EGFR TKIs (27-29). Current EGFR TKIs are targeted to the ATP binding pocket of the enzyme (30).
Specific activating mutations dictate the efficacy of EGFR TKI therapy. For example, in response to EGFR TKI, profound response is observed in tumors that harbor activating EGFR exon 19 deletions and EGFRL858R mutation in exon 21. These tumors also account for almost 90% of all EGFR mutations in NSCLC (31). These mutations result in constitutive activation of EGFR through an increase in ATP binding affinity. On the other hand, a subset of tumors harbor EGFR insertions mutations in exon 20, which does not affect its affinity for ATP and hence a response to EGFR TKIs is uncommon in tumors bearing exon 20 mutations (7,30).
BRAF
BRAF, a member of the Raf kinase family of growth signal transduction protein kinases, is a sentinel component for the MAPK/ERK signaling pathway and regulates cell division, growth and differentiation. In TCGA analysis 3–8% of lung adenocarcinomas harbor somatic alterations in BRAF of which nearly half are BRAFV600E mutation (BRAF class 1 mutation) (1,32). Other common BRAF mutations include the BRAFG469A/V (BRAF class 2 mutation) and BRAFD594G (BRAF class 3 mutation).
BRAF-V600E mutations keep BRAF in its constitutively activated form causing downstream induction of MEK-ERK signaling; and BRAF inhibitors such as vemurafenib and dabrafenib specifically target this constitutive form (33). Furthermore, the addition of a MEK inhibitor in combination of BRAF inhibitors increased the anti-tumor efficacy and have since been FDA approved (34). However, pharmacological inhibitors against less frequent constitutively active dimer (class 2) and RAS driven low activity (class 3) BRAF variants are lacking. Several studies testing inhibitors against class 2 and class 3 BRAF mutant forms are currently ongoing (35,36).
KRAS
KRAS, a small GTPase that belongs to the RAS superfamily, regulates cellular motility, growth and survival in response to trophic and mitogenic stimuli. Growth signaling through activated GTP bound KRAS is mediated through at least three distinct downstream signaling arms-RAF/MAPK, PI3K/AKT and RAL-GEF/RAL signaling pathway (37).
Activating KRAS mutations are present in ~20–30% of patients with NSCLC (1). The most prominent KRAS mutant form is KRAS-G12V—a form that is locked in a constitutively activated GTP bound state. To date, efforts to target KRAS directly have been unsuccessful (38). However, several synthetic lethality screens have identified indirect vulnerability in KRAS mutant lung cancers. For example, in pre-clinical models polo-like kinase 1, RhoA/Rho kinase, nuclear export XPO1 inhibitions have led to selective vulnerability of KRAS mutant lung cancer (39,40) Other potential targets for this strategy include cyclin-dependent kinase 4/6 (CDK4/6) and a phase III trial of the CDK4/6 inhibitor abemaciclib in KRAS-mutant NSCLC is ongoing (41). Recently, in preclinical models, pharmacological inhibition of upstream adapter protein SHP2 has rendered response against KRAS-G12C variant. This variant of RAS can cycle nucleotide to behave in a semi-constitutive manner and hence is responsive to ablation of upstream signaling (42).
ALK and ROS1 rearrangements
Oncogenic ALK and ROS1 gene rearrangements, together, occur in almost 10% of patients with NSCLC (1,43). The resulting degree of overexpression and activation of the fusion protein depends on the nature of the fusion partner. By far the most common fusion partner of ALK is EML4 and of ROS1 is CD74 (2). Four ALK inhibitors that are FDA-approved for use in treating NSCLC are crizotinib (first generation ALK inhibitor), ceritinib, alectinib and brigatinib (second generation ALK inhibitors). Alectinib is now the preferred first-line ALK TKI for treating patients with ALK-rearranged NSCLC, and it resulted in improved outcomes in the ALEX trial (44). Due to the structural homology between the ALK and ROS1 kinase domains, crizotinib, a first-generation drug originally approved for ALK rearranged lung cancers has successfully been applied to ROS1 rearranged cancers as well (45).
Squamous cell lung cancer
FGFR1
In TCGA analysis around 18% of early stage squamous cell carcinoma tumor samples had either copy number amplification or mutations on FGFR1 (12). This observation was in accordance with multiple other earlier studies published on lung cancer cell lines and xenograft models (46,47). Moreover, Zhang and colleagues reported a selective sensitivity of SCLC PDXs against AZD4547, an FGFR inhibitor (48). However, early phase clinical trials testing the FGFR inhibitors, including AZD4547, showed a disappointing response rate ranging from 8–12% (49-51). This poor response rate was attributed to poor correlation between copy number amplification and protein expression as well as co-alterations in PI3K and cell G1/S checkpoint pathways (49).
PI3K/AKT pathway
PI3K/AKT pathway regulates an intricate cell biological signaling network that regulates cell survival, metabolism and proliferation. In TCGA analysis, alterations in PI3K pathway members, such as PTEN and PIK3CA, were seen in much higher frequency in squamous cell lung cancer (52,53). Moreover, PTEN conditional deficiency in mice spontaneously generates squamous like pathophysiology and PTEN and PIK3CA mutations correlate with poor prognosis in human patients (49,54).
However, early phase clinical trials testing BKM120, a PI3K inhibitor, in SCLC patients harboring PIK3CA and PTEN mutations showed no response (55). The failure of the biomarker driven approach was speculatively credited to the presence of co-modifiers and adaptive upregulation of immunosuppressive PD-L1 receptors in tumors (54,56).
CDK4/6
CDK4/6 and their binding partners Cyclin D1-3s are important regulators of mitotic cell division; they are negatively regulated by p53/p21 tumor suppressors. CDK4/6 positively regulate cellular translation potential by causing proteolytic degradation of RB, a negative regulator of E2F (57). During normal conditions G1 to S phase transition is tightly regulated through suppression of E2F transcription factors to avoid aberrant firing of cellular growth programs. In squamous cell carcinoma, CCND1, CyclinD1 encoding gene, is amplified in around 15% cases and CDK4 and 6 are mutated and activated in 45% cases, which indicates that these patients might benefit from targeted CDK4/6 inhibitors (12).
However, in breast and colorectal cancer, the CDK4/6 inhibitor abemaciclib and palbociclib showed at best modest anti-tumor efficacy (58,59). Phase 1 multi-cancer studies of abemaciclib showed a partial response in one (out of 6) patients with squamous cell carcinoma. The patient harbored CDKN2A loss, which is an inhibitor of CDK4. Genomic information for the other patients was not available (60).
The list of identified oncogenic drivers in NSCLC is still expanding. MET and HER2 mutations, RET rearrangements, neurotrophic tyrosine kinase (NTRK) fusions and the loss of neurofibromin 1 (NF1) are some of them. Targeted therapeutic approaches against these alterations are also currently underway (42,61-63).
Small cell lung cancer (SCLC)
p53 and RB1
In TCGA a universal inactivation of p53 and RB1 have been reported in SCLC (12). P53 and RB1 blocks aberrant cell cycle progression by inhibiting of G1 to S phase transition in the absence of mitogenic stimuli. In cancer cells, loss of p53 and RB1 leads to de-repression of cell cycle arrest and evasion of apoptosis, which favors aberrant growth signaling. Hence, in SCLC, inactivation of p53 and RB1 in tumor cells might render them selectively vulnerable against targeted inhibitors against G2/M checkpoints (64). In this regard, CDK4/6 inhibitors are currently being actively pursued (65).
EZH2
EZH2 is a component of the polycomb repressive complex, which methylates histones (H3K27me) to epigenetically silence genes responsible for suppressing cellular growth and division. In SCLC, recurrent overexpression of enhancer of zeste homolog 2 (EZH2) has been reported (66). This overexpression correlates with RB1 loss, as EZH2 is a direct target E2F transcription factors, factors which are sequestered inactive by RB1. Inhibitors against SET domains of EZH2 have been developed by multiple groups resulting in an exciting new opportunity to target non-kinase tumor biomarkers (67). Recent work in patient derived xenografts suggested that combinatorial treatment of EZH2 inhibitors with chemotherapeutic agents blocked chemoresistance (68). Further works on this front will be greatly valuable and is currently undergoing.
FGFR1
FGFR1 is one of the four paralogous transmembrane tyrosine kinases from fibroblast growth factor receptor (FGFR) superfamily that regulates cell proliferation and differentiation via the PI3K/AKT and RAS/RAF/MAPK pathways. In TCGA analysis FGFR1 is amplified in 6% of patients with SCLC (12). However, in preclinical models, FGFR1 protein level, but not copy number was found to be a reliable biomarker for response to FGFR inhibitors (69). Currently, there is a biomarker driven clinical trials for multi kinase inhibitor of FGFRs, ponatinib, ongoing in SCLC patients. Results from this trial might shed light into the reliability of FGFR expression as a biomarker (70).
Notch pathway
In the TCGA analysis the significant alterations in Notch pathway components has been reported in SCLC patients. In preclinical models MEDI0639, a human monoclonal antibody directed against notch pathway ligand DLL4, and OMP-59R5, a human monoclonal antibody targeting Notch 2/3 receptors, inhibits angiogenesis (21,71). Encouraged by this, Rudin and colleagues developed an antibody-drug conjugate Rovalpituzumab teserine (Rova-T) against Notch ligands to deliver cytotoxic payload to Notch high tumors. Phase I clinical trial of Rova-T in patients with high DLL3 expression metastatic SCLC showed a 39% response rate (72). However, severe toxicity for the treatment was also observed. A further study with Rova-T is currently ongoing (73).
MYC
MYC is a transcriptional regulator of aurora kinase A and B, and a known oncogenic driver. Aurora kinases regulate mitotic spindle assembly during mitotic cell division. A myc driven mouse model is a valuable tool for studying hepatocellular carcinoma; however, Myc has recently been implicated in neuroendocrine SCLC oncogenesis (74). SCLC cells with high Myc expression were found to be selectively vulnerable to aurora kinase inhibitors. Clinical trials using orally available aurora kinase inhibitors such as Alisertib are currently ongoing in SCLC (75).
Immunotherapy
Anti-CTLA-4 mAb
CTLA-4 is a surface receptor, expressed usually by immunosuppressive Treg cells. CTLA-4 functions as an immune checkpoint molecule and downregulates cellular immune responses. Cancer cells express CTLA4 complementary CD80/86 molecules to turn on CTLA-4 signaling and downregulate T cell mediated killing of tumors (76). Strategies to block CTLA-4-ligand interaction, using anti-CTLA-4 monoclonal antibody Ipilimumab have shown some efficacy in preclinical models (77). Moreover, a phase 2 trial indicated that ipilimumab increased by 1.1 months the median progression free survival of patients when applied in combination with chemotherapeutic agents (78).
Anti-PD-1 and PD-L1 mAb
Similar to CTLA-4 PD1 and PD-L1 also work as immune checkpoint molecules. Both anti-PD1 and anti-PD-L1 monoclonal antibodies have shown some early success (79,80). Nivolumab was the first PD-1 inhibitor in clinical development and the phase 1 trial using it showed early responses in NSCLC with an overall response rate of 17% (81).
Engineered T cells
Lung cancer is poorly immunogenic, due to their intrinsic inability to present antigens. However, ex vivo engineering of T cell (e.g., chimeric antigen receptor or CAR T cells), to make them better recognize and react to tumor antigens, is an approach being pursued actively. Targets that are highly overexpressed in lung tumors such as MUC1 and mutant EGFR are currently underway for CAR T based therapy (82).
Complete and durable responses to molecular targeted therapy are rare in individuals with advanced-stage solid cancers. Despite a profound response initially, resistance to therapeutic agent occurs through acquired and adaptive processes. Moreover, the presence of co-occurring modifiers that support the so-called driver oncogenes paint a more multi driver and heterogeneous progression model for tumor evolution. Accounting for resistance mechanisms and tumor heterogeneity upfront is the new Achilles’ heel of precision medicine.
Resistance to precision medicine agents
Targeted therapy response often manifests a residual disease state that works as a stepping stone for subsequent tumor progression and acquired resistance. Therefore, there is an urgent need to better understand the molecular basis of residual disease for designing therapeutic strategies to eliminate residual disease.
Resistance to targeted agents can be classified as intrinsic, adaptive resistance and acquired resistance. If a tumor does not respond to the therapy initially; it is classified as intrinsically resistant. Conversely, some tumors respond to the initial treatment, but fail to sustain the efficacy of the therapy due to adaptive and acquired resistance. Adaptive resistance occurs due to a cancer cell’s plastic counter response to the therapy, whereas acquired resistance is governed by either selective propagation of pre-existing or de novo therapy-induced molecular alterations.
Intrinsic resistance
Intrinsically resistant tumors do not respond to targeted therapy. However, often times, combinatorial blockade of intrinsic resistance mechanism act synergistically with the initial targeted therapy. For example, EGFR TKI resistance was implicated in defects in FAS and NF-κB pathway. Interestingly, pharmacological inhibition NF-κB via PBS-1086 overcame this resistance (83). Since this study, multiple other reports have implicated NF-κB signaling in EGFR TKI resistance (84-86).
Acquired and adaptive resistance
Depending on the mechanism of adaptive resistance it can also be sub-classified as on-target or off-target (2). On-target resistance occurs when the primary target of the drug itself is altered, limiting the drug’s ability to inhibit the activity of its target. Off-target resistance occurs through the activation of bypass escape mechanisms that are either parallel or downstream of the target.
Second-site mutation
Resistance can occur via a secondary mutation in the drug target that interferes with inhibition by the targeted therapy. For example, under first generation EGFR TKI treatment, appearance of EGFR-T790M mutation is a classic example of a second site mutation (87). It occurs at a conserved “gatekeeper” threonine residue within the ATP binding pocket and hence limits the first-generation EGFR TKIs ability to bind and inactivate the target. Although, the covalently acting third generation EGFR TKI, osimertinib, overcomes this resistance, another site mutation EGFR-C797S, renders resistance to osimertinib by limiting the drug’s binding (29). Similar second site mutations have been reported for targeted therapy against other oncogenes such as ALK (ALK-L1196M) and ROS1 (ROS1-L2026M) (13,88).
Activation of downstream signaling
Mutational activation of downstream signaling pathway components is another frequent mechanism employed by drug resistant cells to escape upstream signaling blockade. For example, in EGFR driven tumors, resistant cells reactivate MAPK pathway at multiple downstream points so as to render the effect of the drug futile. Resistance to early-generation EGFR TKIs can occur via the acquisition of BRAF mutations (BRAF-G469A or BRAF-V600E) (89). In accordance with this, EGFR TKI resistance also occurs via reactivation of the other two downstream arms of EGFR PI3K/AKT/mTOR and JAK/STAT signaling (90-92).
Activation of bypass signaling
The activation of parallel signaling pathways is another escape mechanism for drug resistant cells. For example, EGFR TKI treatment can activate other members of ERBB family proteins (such as HER2 and HER3) or RTKs required for cell proliferation and survival, thus bypassing inhibition of the original targeted oncogenic driver (93). AXL receptor tyrosine kinase (AXL) is one such RTK that can activate MAPK, PI3K-AKT and NF-κB signaling to promote tumor cell survival and metastasis (94). EGFR TKI resistant patient samples and cell lines had high expression of AXL and its ligand, growth arrest-specific protein 6 (GAS6) (95). AXL inhibitor BGB324 is being evaluated in combination with erlotinib in an ongoing phase I/II study. Interestingly, in ALK and RET driven NSCLC models, AXL overexpression has also been reported as a mechanism of resistance to ALK and RET TKIs indicating a generalized role of AXL for acquired therapy resistance (96). Likewise, EGFR amplification has also been implicated in mechanism of resistance against ALK and RET targeted therapy (97).
Another such generalized bypass mechanism against targeted inhibition occurs via YAP1. YAP is a transcriptional co-activator that serves as an effector for a pathway called Hippo pathway (98). Hippo pathway activation causes proteolytic degradation of YAP leading to attenuation of YAP transcription program (99).
High YAP1 expression was associated with resistance to EGFR TKIs in preclinical models and with poor survival in a cohort of patients with NSCLC and this resistance to EGFR TKIs could be reversed in cell lines by the addition of verteporfin, a small-molecule inhibitor of YAP1 (98,100). Similarly, genetic screening approach found YAP as a mediator for BRAF inhibitor response (101). These observations give rise to an interesting possibility of YAP’s role as a buffer/rheostat between signaling arms of two major driver oncogenes in NSCLC-EGFR and BRAF.
In addition, NF-κB dependent JAK-STAT-IL6 autocrine loop activation was also reported as a bypass mechanism against EGFR TKI treatment. Additionally, this autocrine loop can be blocked using the NF-κB inhibitor PBS-1086. Because the combination therapy using of EGFR TKI and NF-κB inhibitor both enhanced and prolonged the initial response, it exemplifies a prototypic justification for upfront polytherapy (8).
Immunotherapy resistance
Although lung cancer expresses tumor antigens, the tumors are poorly immunogenic as they are ineffective as antigen presenting cells (APC). This gives rise to the intrinsic resistance in majority of lung cancers. Another mechanism of intrinsic resistance is implicated in generic attenuation in T cell intrusion and immunogenicity (e.g., mutation in IFN-γ pathway) (102).
Over time adaptive immunotherapy resistance can also be established through a shift in types of immune cell populations. For example, extensive immunotherapy has been linked to an increase in immunosuppressive Treg and decrease in M2 macrophage population surrounding the tumors. This resulted in secretion of TGF-β and other immunosuppressive cytokines leading to establishment of a paracrine loop (103). Immunotherapy has also been linked to activation of bypass survival signaling such as PI3K/mTOR signaling (56).
Our mechanistic understanding of therapy resistance is much richer than it was a decade ago. However, often times projecting the resistance rout to targeted therapy is difficult due to our primitive understanding of tumor heterogeneity and of the residual disease state.
Tumor heterogeneity
Accumulating evidence suggests the presence of intra- and inter-tumor heterogeneity. Increasingly, it is becoming clear that tumor heterogeneity plays an important role behind the incomplete and short-lived nature of targeted therapy efficacy. Hence, much effort is being put forth to better understand tumor heterogeneity and account for it upfront during strategizing treatment.
The heterogeneity of tumor evolution over time has been described in both advanced-stage EGFR-driven and ALK-driven NSCLC (104,105). Not surprisingly, increased baseline heterogeneity correlated with a shorter duration of response to EGFR TKI therapy (104). Similarly, Blakely et al., through genomic analysis of 1122 EGFR-mutant lung cancer cell free DNA samples, demonstrated that tumor genomic complexity increased over time. Hence, prioritizing therapies that block the more truncal mutations responsible for conferring resistance might hinder branching out and genetic diversification (10). This, in return, would also limit the combination of treatments as more manageable. Analysis from Blakely et al. was able to identify an enrichment of co-occurring mutations on WNT, PI3K and CDK components in the course of tumor evolution. Similar observations were introduced by Gerlinger et al. in primary renal carcinomas and associated metastatic sites, where they demonstrated Darwinian clonal enrichment of mTOR activating mutation in the course of metastasis (106). These combined observations indicate that assaying tumor heterogeneity through longitudinal core and metastatic biopsies coupled with liquid serial biopsy will be informative for risk stratifying patients and assigning them to polytherapy. In parallel, understanding tumor heterogeneity and the trajectory of Darwinian evolution might also enable us to strategize duration of treatments as hypothesized computationally and demonstrated in vitro by Johsson et al. (107).
Discussion
An overarching goal for lung cancer genomics is not only to generate a comprehensive landscape of tumor alterations, but also to map the dynamic evolution of tumors in the presence of genomics-driven therapeutic interventions. The hope is that this, in turn, will lead to novel insights into cancer biology for developing better therapies and prediction models for therapy response and resistance. With the ambitious and large-scale TCGA efforts, which was a deviation from a conventional hypothesis driven approach, we have come close to capturing the snapshot of whole spectrum of cancer alterations (108). However, what remains to be fully understood is how these alterations dynamically evolve through complex interactions among tumor, therapy and microenvironment (109). It is becoming increasingly clear that the conventional single intervention therapies are often inadequate to address the multifactorial aspects of these complex interactions. Hence, the field is increasingly shifting toward a more rational and combinatorial intervention strategy and a comprehensive and genomic understanding of the tumors over time could lead to improved response prediction and therapeutic intervention efficacy, an example of which is “polytherapy” (110-119).
Additionally, ideas to combine different modes of treatments are opening doors for previously untapped opportunities. For example, the prospect of immunotherapy as a consolidation therapy following targeted kinase-inhibitor induction therapy is promising in NSCLC (120). In this regard, the application of engineered CAR T cells that recognize and react against resistance-determining truncal alterations has not been explored yet. These approaches may usher in precise targeting of residual disease-causing tumor and non-tumor cell populations. Additionally, tumors with greater genomic instability may express more neo-antigens and therefore, more susceptible to consolidation immunotherapy. Hence, identifying reliable biomarkers for genomic instability in NSCLC may prove useful not only for treating patients but also for predicting therapy response and risk-stratifying patients.
In this review, we have attempted to summarize recent developments of genomics-guided personalized therapy approaches against lung cancer. Successful implementation of personalized medicine requires comprehensive understanding of the underlying molecular events both at baseline and sequentially throughout the course of therapeutic interventions. We hope that a better genomics level understanding of these events and the evolutionary trajectories that cancers utilize during treatment will yield therapeutic options to forestall tumor evolution and drug resistance and thereby transform aggressive cancers into chronic or curable conditions.
Acknowledgements
Funding: The authors acknowledge funding support from the following sources: NIH/NCI R01CA169338, NIH/NCI R01CA211052, NIH/NCI R01CA204302, NIH/NCI U54CA224081 (each to TG Bivona).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer 2017;17:637-58. [Crossref] [PubMed]
- Liu SV, Giaccone G. Lung cancer: First-line immunotherapy in lung cancer - taking the first step. Nat Rev Clin Oncol 2016;13:595-6. [Crossref] [PubMed]
- Remon J, Besse B, Soria JC. Successes and failures: what did we learn from recent first-line treatment immunotherapy trials in non-small cell lung cancer? BMC Med 2017;15:55. [Crossref] [PubMed]
- Attili I, Passaro A, Pavan A, et al. Combination immunotherapy strategies in advanced non-small cell lung cancer (NSCLC): Does biological rationale meet clinical needs? Crit Rev Oncol Hematol 2017;119:30-9. [Crossref] [PubMed]
- Bivona TG, Doebele RC. A framework for understanding and targeting residual disease in oncogene-driven solid cancers. Nature Med 2016;22:472-8. [Crossref] [PubMed]
- Wu JY, Wu SG, Yang CH, et al. Lung cancer with epidermal growth factor receptor exon 20 mutations is associated with poor gefitinib treatment response. Clin Cancer Res 2008;14:4877-82. [Crossref] [PubMed]
- Blakely CM, Pazarentzos E, Olivas V, et al. NF-kappaB-activating complex engaged in response to EGFR oncogene inhibition drives tumor cell survival and residual disease in lung cancer. Cell Rep 2015;11:98-110. [Crossref] [PubMed]
- Restifo NP, Esquivel F, Kawakami Y, et al. Identification of human cancers deficient in antigen processing. J Exp Med 1993;177:265-72. [Crossref] [PubMed]
- Blakely CM, Watkins TBK, Wu W, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet 2017;49:1693-704. [Crossref] [PubMed]
- Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012;150:1107-20. [Crossref] [PubMed]
- George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47-53. [Crossref] [PubMed]
- Choi YL, Soda M, Yamashita Y, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med 2010;363:1734-9. [Crossref] [PubMed]
- Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 2010;363:1693-703. [Crossref] [PubMed]
- Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer 2010;10:760-74. [Crossref] [PubMed]
- Ding L, Ley TJ, Larson DE, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature 2012;481:506-10. [Crossref] [PubMed]
- Sanchez-Cespedes M, Parrella P, Esteller M, et al. Inactivation of LKB1/STK11 is a common event in adenocarcinomas of the lung. Cancer Res 2002;62:3659-62. [PubMed]
- Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008;455:1069-75. [Crossref] [PubMed]
- Kan SH, Huang F, Tang J, et al. Role of intrapulmonary expression of inducible nitric oxide synthase gene and nuclear factor kappaB activation in severe pancreatitis-associated lung injury. Inflammation 2010;33:287-94. [Crossref] [PubMed]
- Rudin CM, Durinck S, Stawiski EW, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111-6. [Crossref] [PubMed]
- Yen WC, Fischer MM, Axelrod F, et al. Targeting Notch signaling with a Notch2/Notch3 antagonist (tarextumab) inhibits tumor growth and decreases tumor-initiating cell frequency. Clin Cancer Res 2015;21:2084-95. [Crossref] [PubMed]
- Steuer CE, Ramalingam SS. Tumor mutation burden: leading immunotherapy to the era of precision medicine? J Clin Oncol 2018;36:631-2. [Crossref] [PubMed]
- Rizvi H, Sanchez-Vega F, La K, et al. Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) blockade in patients with non-small-cell lung cancer profiled with targeted next-generation sequencing. J Clin Oncol 2018;36:633-41. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Attarian S, Rahman N, Halmos B. Emerging uses of biomarkers in lung cancer management: molecular mechanisms of resistance. Ann Transl Med 2017;5:377. [Crossref] [PubMed]
- Travis WD, Travis LB, Devesa SS. Lung cancer. Cancer 1995;75:191-202. [Crossref] [PubMed]
- Zhou C, Wu YL, Chen G, et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 2011;12:735-42. [Crossref] [PubMed]
- Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol 2013;31:3327-34. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Yasuda H, Park E, Yun CH, et al. Structural, biochemical, and clinical characterization of epidermal growth factor receptor (EGFR) exon 20 insertion mutations in lung cancer. Sci Transl Med 2013;5:216ra177. [Crossref] [PubMed]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol 2001;2:127-37. [Crossref] [PubMed]
- Marchetti A, Felicioni L, Malatesta S, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol 2011;29:3574-9. [Crossref] [PubMed]
- Yao Z, Torres NM, Tao A, et al. BRAF Mutants Evade ERK-dependent feedback by different mechanisms that determine their sensitivity to pharmacologic inhibition. Cancer Cell 2015;28:370-83. [Crossref] [PubMed]
- Planchard D, Besse B, Groen HJM, et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol 2016;17:984-93. [Crossref] [PubMed]
- Yao Z, Yaeger R, Rodrik-Outmezguine VS, et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 2017;548:234-8. [PubMed]
- Dankner M, Rose AAN, Rajkumar S, et al. Classifying BRAF alterations in cancer: new rational therapeutic strategies for actionable mutations. Oncogene 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Rangarajan A, Hong SJ, Gifford A, et al. Species- and cell type-specific requirements for cellular transformation. Cancer Cell 2004;6:171-83. [Crossref] [PubMed]
- McCormick F. KRAS as a therapeutic target. Clin Cancer Res 2015;21:1797-801. [Crossref] [PubMed]
- Kim J, McMillan E, Kim HS, et al. XPO1-dependent nuclear export is a druggable vulnerability in KRAS-mutant lung cancer. Nature 2016;538:114-7. [Crossref] [PubMed]
- Wang J, Hu K, Guo J, et al. Suppression of KRas-mutant cancer through the combined inhibition of KRAS with PLK1 and ROCK. Nat Commun 2016;7:11363. [Crossref] [PubMed]
- Tao Z, Le Blanc JM, Wang C, et al. Coadministration of Trametinib and Palbociclib Radiosensitizes KRAS-mutant non-small cell lung cancers in vitro and in vivo. Clin Cancer Res 2016;22:122-33. [Crossref] [PubMed]
- Nichols RJ, Haderk F, Stahlhut C, et al. Efficacy of SHP2 phosphatase inhibition in cancers with nucleotide-cycling oncogenic RAS, RAS-GTP dependent oncogenic BRAF and NF1 loss. BioRxiv. 2017. Available online: https://www.biorxiv.org/content/early/2017/09/14/188730
- Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007;448:561-6. [Crossref] [PubMed]
- Peters S, Camidge DR, Shaw AT, et al. Alectinib versus Crizotinib in Untreated ALK-positive non-small-cell lung cancer. N Engl J Med 2017;377:829-38. [Crossref] [PubMed]
- Lin JJ, Kennedy E, Sequist LV, et al. Clinical activity of alectinib in advanced RET-rearranged non-small cell lung cancer. J Thorac Oncol 2016;11:2027-32. [Crossref] [PubMed]
- Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med 2010;2:62ra93. [Crossref] [PubMed]
- Kim Y, Hammerman PS, Kim J, et al. Integrative and comparative genomic analysis of lung squamous cell carcinomas in East Asian patients. J Clin Oncol 2014;32:121-8. [Crossref] [PubMed]
- Zhang J, Zhang L, Su X, et al. Translating the therapeutic potential of AZD4547 in FGFR1-amplified non-small cell lung cancer through the use of patient-derived tumor xenograft models. Clin Cancer Res 2012;18:6658-67. [Crossref] [PubMed]
- Paik PK, Shen R, Berger MF, et al. A Phase Ib Open-Label Multicenter Study of AZD4547 in patients with advanced squamous cell lung cancers. Clin Cancer Res 2017;23:5366-73. [Crossref] [PubMed]
- Nogova L, Sequist LV, Perez Garcia JM, et al. Evaluation of BGJ398, a Fibroblast Growth Factor Receptor 1-3 kinase inhibitor, in patients with advanced solid tumors harboring genetic alterations in fibroblast growth factor receptors: results of a global phase i, dose-escalation and dose-expansion study. J Clin Oncol 2017;35:157-65. [Crossref] [PubMed]
- Lim SH, Sun JM, Choi YL, et al. Efficacy and safety of dovitinib in pretreated patients with advanced squamous non-small cell lung cancer with FGFR1 amplification: A single-arm, phase 2 study. Cancer 2016;122:3024-31. [Crossref] [PubMed]
- Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature 2010;463:191-6. [Crossref] [PubMed]
- Jin G, Kim MJ, Jeon HS, et al. PTEN mutations and relationship to EGFR, ERBB2, KRAS, and TP53 mutations in non-small cell lung cancers. Lung Cancer 2010;69:279-83. [Crossref] [PubMed]
- Xu C, Fillmore CM, Koyama S, et al. Loss of Lkb1 and Pten leads to lung squamous cell carcinoma with elevated PD-L1 expression. Cancer Cell 2014;25:590-604. [Crossref] [PubMed]
- Vansteenkiste JF, Canon JL, De Braud F, et al. Safety and Efficacy of Buparlisib (BKM120) in Patients with PI3K pathway-activated non-small cell lung cancer: results from the phase II BASALT-1 Study. J Thorac Oncol 2015;10:1319-27. [Crossref] [PubMed]
- Lastwika KJ, Wilson W 3rd, Li QK, et al. Control of PD-L1 Expression by Oncogenic Activation of the AKT-mTOR pathway in non-small cell lung cancer. Cancer Res 2016;76:227-38. [Crossref] [PubMed]
- Dickson MA, Schwartz GK. Development of cell-cycle inhibitors for cancer therapy. Curr Oncol 2009;16:36-43. [PubMed]
- Gelbert LM, Cai S, Lin X, et al. Preclinical characterization of the CDK4/6 inhibitor LY2835219: in-vivo cell cycle-dependent/independent anti-tumor activities alone/in combination with gemcitabine. Invest New Drugs 2014;32:825-37. [Crossref] [PubMed]
- Fry DW, Harvey PJ, Keller PR, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther 2004;3:1427-38. [PubMed]
- Patnaik A, Rosen LS, Tolaney SM, et al. Efficacy and Safety of Abemaciclib, an Inhibitor of CDK4 and CDK6, for patients with breast cancer, non-small cell lung cancer, and other solid tumors. Cancer Discov 2016;6:740-53. [Crossref] [PubMed]
- Farago AF, Le LP, Zheng Z, et al. Durable Clinical Response to Entrectinib in NTRK1-rearranged non-small cell lung cancer. J Thorac Oncol 2015;10:1670-4. [Crossref] [PubMed]
- Kobayashi Y, Mitsudomi T. Not all epidermal growth factor receptor mutations in lung cancer are created equal: Perspectives for individualized treatment strategy. Cancer Sci 2016;107:1179-86. [Crossref] [PubMed]
- Huang Q, Schneeberger VE, Luetteke N, et al. Preclinical Modeling of KIF5B-RET Fusion Lung Adenocarcinoma. Mol Cancer Ther 2016;15:2521-9. [Crossref] [PubMed]
- Dobbelstein M, Sorensen CS. Exploiting replicative stress to treat cancer. Nat Rev Drug Discov 2015;14:405-23. [Crossref] [PubMed]
- A Study of Abemaciclib (LY2835219) in Participants With Previously Treated KRAS Mutated Lung Cancer (JUNIPER). US National Library of Medicine ClinicalTrialsgov, 2017.
- Hubaux R, Thu KL, Coe BP, et al. EZH2 promotes E2F-driven SCLC tumorigenesis through modulation of apoptosis and cell-cycle regulation. J Thorac Oncol 2013;8:1102-6. [Crossref] [PubMed]
- Tan J, Yang X, Zhuang L, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev 2007;21:1050-63. [Crossref] [PubMed]
- Gardner EE, Lok BH, Schneeberger VE, et al. Chemosensitive relapse in small cell lung cancer proceeds through an EZH2-SLFN11 Axis. Cancer Cell 2017;31:286-99. [Crossref] [PubMed]
- Wynes MW, Hinz TK, Gao D, et al. FGFR1 mRNA and protein expression, not gene copy number, predict FGFR TKI sensitivity across all lung cancer histologies. Clin Cancer Res 2014;20:3299-309. [Crossref] [PubMed]
- Study of Ponatinib in Patients With Lung Cancer Preselected Using Different Candidate Predictive Biomarkers. US National Library of Medicine ClinicalTrialsgov, 2014.
- Jenkins DW, Ross S, Veldman-Jones M, et al. MEDI0639: a novel therapeutic antibody targeting Dll4 modulates endothelial cell function and angiogenesis in vivo. Mol Cancer Ther 2012;11:1650-60. [Crossref] [PubMed]
- Rudin CM, Pietanza MC, Bauer TM, et al. Rovalpituzumab tesirine, a DLL3-targeted antibody-drug conjugate, in recurrent small-cell lung cancer: a first-in-human, first-in-class, open-label, phase 1 study. Lancet Oncol 2017;18:42-51. [Crossref] [PubMed]
- Rovalpituzumab Tesirine in Delta-Like Protein 3-Expressing Advanced Solid Tumors. US National Library of Medicine ClinicalTrialsgov. 2016.
- Mollaoglu G, Guthrie MR, Bohm S, et al. MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to aurora kinase inhibition. Cancer Cell 2017;31:270-85. [Crossref] [PubMed]
- Phase 2 Study of Alisertib (MLN8237) in Combination With Paclitaxel Versus Placebo in Combination With Paclitaxel as Second Line Therapy for Small Cell Lung Cancer (SCLC). US National Library of Medicine ClinicalTrialsgov, 2014.
- Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature 2011;480:480-9. [Crossref] [PubMed]
- Di Giacomo AM, Biagioli M, Maio M. The emerging toxicity profiles of anti-CTLA-4 antibodies across clinical indications. Semin Oncol 2010;37:499-507. [Crossref] [PubMed]
- The Addition of Ipilimumab to Carboplatin and Etoposide Chemotherapy for Extensive Stage Small Cell Lung Cancer (ICE). US National Library of Medicine ClinicalTrialsgov, 2011.
- Fehrenbacher L, Spira A, Ballinger M, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 2016;387:1837-46. [Crossref] [PubMed]
- Garon EB, Rizvi NA, Hui R, et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 2015;372:2018-28. [Crossref] [PubMed]
- Topalian SL, Hodi FS, Brahmer JR, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012;366:2443-54. [Crossref] [PubMed]
- Feng K, Guo Y, Dai H, et al. Chimeric antigen receptor-modified T cells for the immunotherapy of patients with EGFR-expressing advanced relapsed/refractory non-small cell lung cancer. Sci China Life Sci 2016;59:468-79. [Crossref] [PubMed]
- Bivona TG, Hieronymus H, Parker J, et al. FAS and NF-kappaB signalling modulate dependence of lung cancers on mutant EGFR. Nature 2011;471:523-6. [Crossref] [PubMed]
- Galvani E, Sun J, Leon LG, et al. NF-kappaB drives acquired resistance to a novel mutant-selective EGFR inhibitor. Oncotarget 2015;6:42717-32. [Crossref] [PubMed]
- Li L, Gu X, Yue J, et al. Acquisition of EGFR TKI resistance and EMT phenotype is linked with activation of IGF1R/NF-kappaB pathway in EGFR-mutant NSCLC. Oncotarget 2017;8:92240-53. [PubMed]
- Chiu CF, Chang YW, Kuo KT, et al. NF-kappaB-driven suppression of FOXO3a contributes to EGFR mutation-independent gefitinib resistance. Proc Natl Acad Sci U S A 2016;113:E2526-35. [Crossref] [PubMed]
- Lin L, Asthana S, Chan E, et al. Mapping the molecular determinants of BRAF oncogene dependence in human lung cancer. Proc Natl Acad Sci U S A 2014;111:E748-57. [Crossref] [PubMed]
- Casaluce F, Sgambato A, Sacco PC, et al. Resistance to Crizotinib in Advanced Non-Small Cell Lung Cancer (NSCLC) with ALK Rearrangement: Mechanisms, Treatment Strategies and New Targeted Therapies. Curr Clin Pharmacol 2016;11:77-87. [Crossref] [PubMed]
- Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A 2008;105:2070-5. [Crossref] [PubMed]
- Gao SP, Chang Q, Mao N, et al. JAK2 inhibition sensitizes resistant EGFR-mutant lung adenocarcinoma to tyrosine kinase inhibitors. Sci Signal 2016;9:ra33. [Crossref] [PubMed]
- Chaib I, Karachaliou N, Pilotto S, et al. Co-activation of STAT3 and YES-Associated Protein 1 (YAP1) Pathway in EGFR-Mutant NSCLC. J Natl Cancer Inst 2017.109. [PubMed]
- Jacobsen K, Bertran-Alamillo J, Molina MA, et al. Convergent Akt activation drives acquired EGFR inhibitor resistance in lung cancer. Nat Commun 2017;8:410. [Crossref] [PubMed]
- Arteaga CL, Engelman JA. ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell 2014;25:282-303. [Crossref] [PubMed]
- Scaltriti M, Elkabets M, Baselga J. Molecular Pathways: AXL, a Membrane Receptor Mediator of Resistance to Therapy. Clin Cancer Res 2016;22:1313-7. [Crossref] [PubMed]
- Zhang Z, Lee JC, Lin L, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet 2012;44:852-60. [Crossref] [PubMed]
- Yamaguchi N, Lucena-Araujo AR, Nakayama S, et al. Dual ALK and EGFR inhibition targets a mechanism of acquired resistance to the tyrosine kinase inhibitor crizotinib in ALK rearranged lung cancer. Lung Cancer 2014;83:37-43. [Crossref] [PubMed]
- Katayama R, Shaw AT, Khan TM, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung Cancers. Sci Transl Med 2012;4:120ra17. [Crossref] [PubMed]
- Lin L, Bivona TG. The Hippo effector YAP regulates the response of cancer cells to MAPK pathway inhibitors. Mol Cell Oncol 2015;3:e1021441. [Crossref] [PubMed]
- Yu FX, Zhao B, Guan KL. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 2015;163:811-28. [Crossref] [PubMed]
- Lee JE, Park HS, Lee D, et al. Hippo pathway effector YAP inhibition restores the sensitivity of EGFR-TKI in lung adenocarcinoma having primary or acquired EGFR-TKI resistance. Biochem Biophys Res Commun 2016;474:154-60. [Crossref] [PubMed]
- Lin L, Sabnis AJ, Chan E, et al. The Hippo effector YAP promotes resistance to RAF- and MEK-targeted cancer therapies. Nat Genet 2015;47:250-6. [Crossref] [PubMed]
- Sharma P, Hu-Lieskovan S, Wargo JA, et al. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 2017;168:707-23. [Crossref] [PubMed]
- Woo EY, Yeh H, Chu CS, et al. Cutting edge: Regulatory T cells from lung cancer patients directly inhibit autologous T cell proliferation. J Immunol 2002;168:4272-6. [Crossref] [PubMed]
- Suda K, Murakami I, Sakai K, et al. Heterogeneity in resistance mechanisms causes shorter duration of epidermal growth factor receptor kinase inhibitor treatment in lung cancer. Lung Cancer 2016;91:36-40. [Crossref] [PubMed]
- Gainor JF, Dardaei L, Yoda S, et al. Molecular mechanisms of resistance to first- and second-generation ALK Inhibitors in ALK-rearranged lung cancer. Cancer Discov 2016;6:1118-33. [Crossref] [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [Crossref] [PubMed]
- Jonsson VD, Blakely CM, Lin L, et al. Novel computational method for predicting polytherapy switching strategies to overcome tumor heterogeneity and evolution. Sci Rep 2017;7:44206. [Crossref] [PubMed]
- The future of cancer genomics. Nat Med 2015;21:99. [Crossref] [PubMed]
- Riaz N, Havel JJ, Makarov V, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 2017;171:934-49.e15. [Crossref] [PubMed]
- Hrustanovic G, Olivas V, Pazarentzos E, et al. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nat Med 2015;21:1038-47. [Crossref] [PubMed]
- Govindan R, Mandrekar SJ, Gerber DE, et al. ALCHEMIST Trials: A golden opportunity to transform outcomes in early-stage non-small cell lung cancer. Clin Cancer Res 2015;21:5439-44. [Crossref] [PubMed]
- Jänne PA, Cohen RB, Laird AD, et al. Phase I safety and pharmacokinetic study of the PI3K/mTOR inhibitor SAR245409 (XL765) in combination with erlotinib in patients with advanced solid tumors. J Thorac Oncol 2014;9:316-23. [Crossref] [PubMed]
- Price KA, Azzoli CG, Krug LM, et al. Phase II trial of gefitinib and everolimus in advanced non-small cell lung cancer. J Thorac Oncol 2010;5:1623-9. [Crossref] [PubMed]
- Spigel DR, Ervin TJ, Ramlau RA, et al. Randomized phase II trial of Onartuzumab in combination with erlotinib in patients with advanced non-small-cell lung cancer. J Clin Oncol 2013;31:4105-14. [Crossref] [PubMed]
- Nathan SD, Corris PA. Upfront combination therapy: does the AMBITION study herald a new era in the treatment of pulmonary arterial hypertension? Thorax 2016;71:107-9. [Crossref] [PubMed]
- Rosell R, Dafni U, Felip E, et al. Erlotinib and bevacizumab in patients with advanced non-small-cell lung cancer and activating EGFR mutations (BELIEF): an international, multicentre, single-arm, phase 2 trial. Lancet Respir Med 2017;5:435-44. [Crossref] [PubMed]
- Moran T, Felip E, Keedy V, et al. Activity of dalotuzumab, a selective anti-IGF1R antibody, in combination with erlotinib in unselected patients with Non-small-cell lung cancer: a phase I/II randomized trial. Exp Hematol Oncol 2014;3:26. [Crossref] [PubMed]
- Jänne PA, Shaw AT, Camidge DR, et al. Combined Pan-HER and ALK/ROS1/MET inhibition with dacomitinib and crizotinib in advanced non-small cell lung cancer: results of a phase I Study. J Thorac Oncol 2016;11:737-47. [Crossref] [PubMed]
- Miyajima N, Tsutsumi S, Sourbier C, et al. The HSP90 inhibitor ganetespib synergizes with the MET kinase inhibitor crizotinib in both crizotinib-sensitive and -resistant MET-driven tumor models. Cancer Res 2013;73:7022-33. [Crossref] [PubMed]
- Berman AT, Simone CB 2nd. Immunotherapy in locally-advanced non-small cell lung cancer: releasing the brakes on consolidation? Transl Lung Cancer Res 2016;5:138-42. [PubMed]


