Chromatin remodeling defects in pediatric brain tumors
Introduction
Pediatric brain tumors are regarded as the most prevalent solid neoplasms and the principal reason of death in childhood. High grade gliomas (HGGs) are rare (8%), but account for the most lethal brain cancer in children (1). Despite the aggressive treatments which include combination of neurosurgery, chemotherapy and radiation therapy in most cases the outcome is poor (2). The 2-year survival for children with diffuse intrinsic pontine gliomas (DIPGs) is less than 10% and for those with supratentorial (ST) HGGs doesn’t exceed 30% (3).
Low grade gliomas (LGGs) are the most frequent tumors of the central nervous system (CNS) that appear in childhood. LGGs are very heterogeneous, formed within the brainstem, brain, and spinal cord, with distinctive characteristics, occurrence patterns, treatment options and responses and variable survival rates (4). Current treatment includes surgery, chemotherapy, radiation and their combination (4). Patients’ survival can vary with a 5-year overall survival in pilocytic astrocytomas (PAs) being described as high as 100%, while this percentage appears to be lower than 50% in diffuse fibrillary astrocytomas (DA) (5).
The most prevalent malignant pediatric brain tumor is medulloblastoma (MB) appertain to embryonal tumors. MB is a brain tumor with great heterogeneity and the second-most frequent brain cancer in children following PAs (6).
Ependymoma (EPN) also displays clinical and genetic heterogeneity with most EPs being infratentorial tumors, arising in or around the fourth ventricle and constituting approximately 6% to 10% of all pediatric brain tumors (7). Surgical resection is the first line treatment, since EPs are rarely metastatic at diagnosis or recurrence.
Recently, efforts have been made to elucidate the genetic and epigenetic mechanisms of pediatric brain neoplasms (8). Deregulation of epigenetic mechanisms may provoke aberrant gene expression, contributing to tumor formation (9). The elucidation of these alterations is crucial for prognosis and the possibility to revert these changes has been valuable for drug development and for improvement of current tumor-specific therapeutic strategies (8).
Epigenetic mechanisms in tumor development
Epigenetics refer to studies of mitotically heritable modifications in gene expression that do not involve changes in DNA sequence, presenting a link between genotype and phenotype. In DNA replication the paternal epigenetic information is translated by enzymes known as “readers” which lead to the recruitment of other proteins termed as “writers”, or “erasers” and preserve this information in the daughter strands (10). The main epigenetic mechanisms are DNA methylation, histone modifications and small non-coding RNAs (miRNAs).
The most well-studied epigenetic mechanism is DNA methylation, being essential for the expression of critical genes as well as for chromatin remodeling (8). DNA methylation involves addition of a methyl group at the 5’ position of the pyrimidine ring of cytosine in cytosine-phosphate-guanine (CpG) dinucleotides and is commonly related to transcriptional silencing (8). CpG nucleotides are frequently found at the 5’ end of the gene promoter and/or the first exon region as big clusters, termed CpG islands (8). The CpG nucleotides inside CpG islands are usually unmethylated, but more than 80% of those traced outside of CpG islands are commonly found methylated (10). DNA methyltransferases (DNMTs) are catalyzing the reaction of methylation and can be distinguished into two subgroups: maintenance methylation and de novo methylation (10). The maintenance methyltransferase DNMT1 is responsible for restoration of the parental DNA methylation pattern. De novo methylation is essential during embryogenesis and cell development, being mediated by DNMT3a and DNMT3b functions (10).
Posttranslational modifications are targeting the amino-terminal of histones in a dynamic and reversible manner under the control of “histone code” which regulates the various combinations of histone modifications that modulate the genetic information of the transcript (10). Histone methylation is a covalent chemical modification that involves the addition of a methyl group on a lysine or arginine residue catalyzed by histone methyltransferases (HMTs). Several histone methylations have been identified, including the lysine (K) residues of histone H3 (K4, K9, K27, K36 and K79) and histone H4 (K20), and the arginine (R) residues of histone H3 (R2, R17 and R26) and histone H4 (R3) (10). Methylations of H3K4, H3K36, and H3K79 are linked to transcriptional activation, whereas methylations of H3K9, H3K27, and H4K20 are accompanied by gene repression (10). Another critical histone modification is acetylation which involves the relocation of an acetyl group from acetyl-CoA to the lysine ε-amino groups on the N-terminal of histone tails by histone acetyltransferases (HATs) (10). Histone acetylation is responsible for the interaction between the negatively charged DNA and histones, as a result of an increase in negative charge (8). This modification is commonly associated with the open state of chromatin, the accessibility of DNA to the binding proteins, and increased transcriptional activity. The reverse reaction, deacetylation, is triggered by histone deacetylases (HDACs) which remove the acetyl groups from histones contributing to chromatin condensation and transcriptional repression (10). HDACs are implicated in several signaling pathways and they are responsible for repressive chromatin complexes. Several HDAC inhibitors that result in chromatin decompression and gene activation have been identified and investigated for their therapeutic value in neurodegenerative disorders and cancer (10).
RNA-based mechanisms consist of small non-coding oligonucleotides, microRNAs (miRNAs) and short interfering RNAs (siRNAs) that contribute to gene silencing (10). By contrast to messenger RNAs (mRNAs), miRNAs are biologically stable and are not easily decomposed by RNAses due to their incorporation into the miRNA-containing RNA-induced silencing complex (RISC) (10). This contributes in RNA silencing and post-transcriptional regulation of gene expression through translational repression or mRNA cleavage, by guiding RISC to detect messenger RNAs (10). MicroRNAs are able to regulate the translation and mRNA transcripts in the cytoplasm through binding to the 3’ untranslated region (10). MicroRNA expression can be modulated by promoter methylation or acetylation of histones (10). Short interfering RNAs are acting both in the cytoplasm and nucleus, mediating chromatin-dependent and posttranscriptional gene silencing (10). The contribution of miRNA in tumorigenesis has been revealed recently with their expression being associated with apoptosis and cell cycle arrest (11). Furthermore, miRNAs implicated in metastasis and invasion have been found upregulated in tumors (11). Recent progress in the elucidation of mechanisms by which miRNAs and siRNAs regulate gene expression have endorsed the design and synthesis of therapeutically effective molecules that induce silencing of target genes in vivo (10).
The epigenetic marks are generated on genomic DNA during development and differentiation (10). These modifications establish active and repressive chromatin structures and operate as molecular switches turning gene expression either “on” or “off”, or by altering overall gene expression levels (8).
Genetic landscape of gliomas
The majority of pediatric LGGs (pLGGs) present alterations in the RAS/MAP kinase pathway. The most frequent change is a fusion of B-Raf proto-oncogene (BRAF) with KIAA1549, which induces constant activation of BRAF kinase domain resulting in the enhancement of mitogen-activated protein kinase (MAPK) pathway (12). In a small percentage of pLGGs, a missense mutation of BRAFV600E has been identified (13). Another important contributor to the development of pLGGs involves mutations on the phosphatidylinositol 3-kinase (PI3K)/mammalian target of rapamycin (mTOR) pathway, affecting cell proliferation and growth (14,15). Several components of this pathway are overexpressed in pLGGs and have been linked to poor prognosis (16). Other frequently altered genes include fibroblast growth factor receptor 1 (FGFR1), tumor Protein p53 (TP53) and neurofibromatosis 1 (NF1), and they were detected in 17.6% (22/125), 5.6% (7/125) and 8.8% (11/125) of cases, respectively (17). Mutations on NF1 cause activation of platelet-activating factor (PAF) pathway that leads to tumorigenesis (18).
According to Sturm et al. there are six subgroups of HGG based on DNA methylation patterns, gene-expression profiles and mutation profiles, isocitrate dehydrogenase (IDH) mutations, platelet-derived growth factor receptor alpha amplification (PDGFRA), receptor tyrosine kinase I, II (RTK I, II), lysine 27 (K27), glycine 34 (G34), and mesenchymal [very few copy number alterations (CNAs) or point mutations] (19).
PDGFRA amplifications are frequently detected in a proportion of pHGGs and possess a key role in tumor development by contrast to mutations in IDH that are absent on pediatric gliomas (20). A large amount of pediatric HGG is characterized by mutations in genes that encode histone 3, in particularly H3 Histone family member 3A (H3F3A) or H3.1 genes Histone cluster 1 H3 family member B (HIST1H3B). These missense mutations result in amino acid substitutions that generate two mutants: K27M mutant, where lysine of position 27 is replaced by methionine, and G34R or G34V mutant, where either arginine or valine substitutes glycine at position 34 (21,22).
H3.3K27M is spreading throughout the midline and pons, and occurs in 63% of DIPG and in 59.7% of non-brainstem midline tumors. In all locations, these mutations were associated with a considerably shorter survival period (overall median 11 months, 2-year overall survival 4.7%). H3.1/3.2K27M was highly specific to the pons (21.4%) representative of a younger age group (median 5.0 years) with a significantly longer overall survival (median 15.0 months) than H3.3K27M (23).
Moreover, these mutations affect other mutations, for example the mutation H3.3K27M observed in the pons is correlated with TP53 loss of function mutations (60%) and PDGFRA gain-of-function mutations or amplifications (40%) (21,24), while the H3.1K27M mutations in the pons are associated with recurrent gain of function somatic mutations in the activin A receptor type 1 (ACVR1; 20%) (25,26). As for the CNAs the most frequent chromosomal changes found in children are chromosome 1q gain (20%), loss of 16q (18%), and loss of 4q (15%) (27). Common large-scale chromosomal alterations include chromosome 17 and 9. More specifically the loss of 17p is targeting TP53 and is linked to shorter survival independently of the subgroup or the tumor location. The gain of 9q34 has been associated with shorter overall survival in pHGGs and DIPGs (23).
Epigenetic histone modifications in gliomas
In pHGGs, there are two mutations implicated in regulatory posttranslational modifications, G34V/R and K27M (21). The role of histone lysine methylation is crucial for gene expression and the state of chromatin (28).
G34V/R mutation occurs in H3.3 histone tail and is reported to correlate with reduced H3K36 methylation through loss of function mutations in the N-methyltransferase SET domain-containing 2 (SETD2) (29). This reduction is associated with enhancement of gene expression. Mutations in SETD2 methyltransferase are taking place solely in hemispheric HGGs, and are more frequently observed in pediatric patients (15% of tumors) than adults (8% of tumors) (29). These missense mutations were not detected in LGGs or midline structures. SETD2 encodes the histone H3K36 tri-methyltransferase in humans and affects the function of histone H3.3. The location of tumor origin in gliomas may be affected by these mutations (30). Additionally, the oncogene MYCN was upregulated by G34R/V mutation and led to transcriptional upregulation through altered genomic binding of methylated H3K36 to specific gene loci (31,32). MYCN was found to be implicated in many cancers including glioblastoma in mouse (31,33).
Except from G34V/R, another frequent posttranslational histone modification is the methylation or acetylation of lysine 27 (K27) in all histone 3 variants (24). The histone N-methyltransferase, enhancer of zeste homologue 2 (EZH2) catalyzes the mono-, di-, or trimethylation of K27 and induces gene silencing. On the other hand, acetylation of K27 leads to activation of gene transcription and gene expression (34). Sometimes due to K27M mutation, there is a reduction of H3K27me2/3 which causes transcriptional activation. However, K27 mutation may also increase H3K27me3 leading to silencing of tumor suppressor gene expression (35).
The polycomb repressive complex 1 (PRC1) and polycomb repressive complex 2 (PRC2) compose the polycomb-group of proteins (PcG). PRC2 complex acts as a HMT through its subunits EZH1 and EZH2 and trimethylates histone H3 on lysine 27 (H3K27me3) resulting in gene silencing (36). A study by Lewis et al. reported the inhibition of PRC2 as a result of abnormal binding of K27M to EZH2 (37). This aberrant remodeling of EZH2 is causing reduction of H3K27 methylation and is leading to gene activation and DNA hypomethylation (37,38). The result of DNA hypomethylation is gene activation and cell differentiation due to CpG hypomethylator phenotype (39). Although, there is a decrease of H3K27 methylation in K27M mutated gliomas, another study reported enhancement of HMTs EZH2 and H3K27 methylation at defined gene loci (40).
Genetic landscape of MBs
In 2012, four MB subgroups were established with distinct clinical, pathological and molecular features, namely Wingless (Wnt), sonic hedgehog (SHH), group 3 and group 4. However, more recent publications propose a different classification encompassing more subgroups and a biological overlap between group 3 and 4 (41).
Wnt tumors may be the rarest (11%) subgroup of MB but it is well-studied with really good prognosis. Wnt subgroup is named after the predominant activation of Wingless signaling pathway (Wnt) that involves a family of growth factor receptors with function during embryonic development (42). Molecular analysis of Wnt MBs shows that the most frequent mutations occur on the gene encoding for β-catenin (43). These somatic catenin beta 1 (CTNNB1) mutations were found in 85% of the patients. Another characteristic alteration found on patient with Wnt MB is monosomy 6 (83%) (44).
SHH tumors account for almost the 30% of all MBs (45). They have an intermediate prognosis ranking between good prognosis of Wnt tumors and decreased prognosis of group 3 tumors. They have been correlated with mutations in Patched 1 (PTCH1), but also in smoothened (SMO) and suppressor of fused (SUFU) genes leading to over activation of the sonic hedgehog signaling (SHH) pathway (43). Additionally, SHH tumors exhibit increased N-myc proto-oncogene (MYCN) expression levels (46).
Group 3 MB is characterized as classic MBs with dismal prognosis and frequently metastatic, harboring elevated MYC expression (46).
Group 4 are the most frequent MBs (34%) and even though they are often metastatic, they are correlated with an intermediate prognosis, similar to SHH tumors. The majority of this group harbors an isochromosome 17q. Group 4 MBs are also associated with MYCN and cyclin dependent kinase 6 (CDK6) amplification but minimal MYC over-expression (43).
Epigenetic histone modifications in MBs
In MBs, most common mutations affect genes that target histone methylation (47). More specifically, molecular screening of MBs revealed truncating mutations in histone lysine methyltransferases MLL3/KMT2C and MLL2/KMT2D that are responsible for the histone methylation H3K4me2/3, associated with “open” chromatin and gene expression (48). These mutations are suppressing tumor formation and are found in 16% of MB as well as in SHH and group 4 (48). Moreover, mutations in genes encoding for methyltransferases SET and MYND domain containing 4 (SMYD4) and euchromatic histone lysine methyltransferase 1 (EHMT1), acetyltransferase MYST3, demethylases JMJD2B and JMJD2C and several PcG have been associated with hypomethylation of H3K9 (49). Furthermore, mutations in trithorax group genes, lysine demethylase 6A (KDM6A) and lysine demethylase 6B (KDM6B), KDM6A (UTX) encoded by lysine demethylases (KDM) result in elevation of H3K27me3 and are commonly found in G4-MB (47) along with overexpression of the PcG protein, EZH2 which is involved in stem cell maintenance by repressing lineage differentiation genes (47).
Apart of lysine methyltransferases, lysine acetyltransferases (HATs) are also involved in brain development and they are linked to poor survival such as the downregulation of H4K16 HAT, hMOF (50). SHH tumors are overexpressing somatic modifications that target HATs (44). In contrast, HDACs are associated with gene silencing and some of them (HDAC5 and HDAC9) are upregulated in MB leading to deregulated cell cycle (51).
In group 3 and 4 modifications in histone demethylases have been detected that induce aberrant histone methylation of H3K4 and H3K27 (48). Bromodomain (BRD) and extra-terminal (BET)-containing proteins (BET/BRD) are binding in acetylated histones and are responsible to manage MYC levels in group 3 MB (52). In recent preclinical studies BET inhibitor (JQ1) is evaluated for its therapeutic role in MBs (53).
Although elucidation of the mechanism that histone modifications mediate pathogenesis of MB is still missing, some evidence suggests that their net effect is to shift the balance between H3K27 and H3K4 methylation, resulting in silencing of genes responsible for progenitor cell differentiation (54-56). Suppressing H3K27me3 by silencing the home box gene Orthodenticle Homeobox 2 (OTX2) leads to a differentiated phenotype in MBs (57,58). Whether the presence of “repressive” heterochromatin (high H3K27me3 and low H3K4me3) in group 3 and group 4 MBs reflects the epigenetic status of the tumor-initiating/propagating cells that generate these tumors, or whether it is a consequence of mutations in genes that regulate these modifications, requires further investigation (47).
Genetic landscape of EPNs
In 2016, the World Health Organization (WHO) classified ependymal tumors into five main subtypes: subependymoma, myxopapillary EPN, EPN, RELA fusion-positive, and anaplastic EPN (7).
The majority of pediatric ST EPNs exhibits gene fusions involving RELA. Chromosome 11 open reading frame 95 (C11orf95-RELA) fusion is associated with constitutive induction of the nuclear factor-kappa B (NF-κB) pathway by an increase in a RELA-encoded transcription factor p65 (59).
Another molecular subgroup for ST EPN is characterized by the fusion of yes-associated protein 1 (YAP1), a transcriptional regulator involved in proliferation, with other genes such as mastermind like domain containing 1 (MAMLD1) and family with sequence similarity 118 member B (FAM118B) (59).
Epigenetic histone modifications in EPNs
Recent studies unraveling the epigenetic pattern of EPN with particular emphasis on histone modifications identified a decrease in H3K27me3 in ~80% of posterior fossa (PF) EPN (60). PF-ve EPN are not characterized by recurrent genetic mutations by contrast to H3K27M gliomas (59,61). Several studies suggest that there is a positive association between unmethylated CpG islands and PRC2 recruitment which depends on DNA methylation (62,63). By contrast, in PF EPN, as a result of the increased CpGi methylation, H3K27me3 is reduced due to incapability of PRC2 to access chromatin (60). Additionally, Bayliss et al. reported that the PF radial glia displayed reduced H3K27me3 through early development, reflecting which tumor formation is going to be developed (60).
Finally, H3K27M gliomas and PF-ve/PFA EPN share many similarities such as tumor location and patient age, and perhaps this chromatin state defined by DNA and H3K27 hypomethylation could have a crucial role in transformation and/or growth of tumor (60). The elucidation of these regulators that share this chromatin state may be critical in understanding the biology of PF EPN.
Biomarker potential of histone alterations in pediatric brain neoplasms
The last decade has seen major advances in pediatric neuro-oncology. Genomic and epigenomic analyses have underlined the heterogeneity of HGG, MB and EPN, and recognized some of the key molecular alterations associated with each tumor subtype (47).
Huse et al. applied whole-exome and next-generation sequencing in order to analyze the molecular profile of MBs and HGGs (64). MBs have been classified into four subtypes based on their molecular differences. Wnt tumors which comprise the first category are frequently harboring CTNNB1 mutations. These mutations are affecting Wnt signaling pathway and are correlated with a prognostic and diagnostic role and favorable survival of patients. SHH tumors are frequently characterized by PCTCH1 mutations and are responsible for the activation of SHH pathway, with a potential diagnostic role (65). Additionally, other genetic aberrations are also found indicating the relatively favorable prognosis associated with Wnt-MB and the poor outcomes associated with G3-MB, SHH-MB with TP53 mutation, and PF-EPN-A (47). MYCN amplification may also have a prognostic significance, often correlated with poor survival (65).
In pHGGs, research findings detected chromatin remodeling defects, mainly mutations in histone H3F3A with the replacement of lysine 27 with methionine (K27M) to be found in 78% of DIPGs whereas one-third of non-brainstem HGGs carries K27M or G34V/R mutations (38). These mutations are correlated with poor outcome therefore can be used for prognosis. TP53 and ATRX mutations are commonly found in HGGs but their prognostic value remains unclear (66). PDGFRA mutations which are correlated with poor survival are frequently observed in HGGs and may also have a prognostic role (67).
On the contrary to HGGs, LGGs are mainly characterized by activation of BRAF oncogene. The fusion gene KIAA1549:BRAF is found in 50–70% of PAs and is generated by tandem duplication at 7q34 deregulating the MAPK signaling pathway, affecting cell proliferation, differentiation and apoptosis (65).
Some studies showed that this fusion gene could improve patients’ survival but it is still uncertain if it useful in prognosis. Furthermore, the frequent BRAF modifications in PAs may have a diagnostic role in differentiating PAs from grade II gliomas. However, BRAF V600E mutations are found both in LGGs and in HGGs with partial diagnostic and predictive value (65).
In EPN, several genes such as JHDMD1, ASAH1 GNAO1, IMMT, IPO7 and CISD3 are found implicated in cell proliferation, signaling pathways, methylation and tumor development. These genes could have a potent prognostic role in EPN because they are participating in deregulation of cell cycle, in tumor formation, cell migration and possibly in metastasis (68).
Except of the molecular profiling of pediatric brain neoplasms, epigenetic changes are also critical during their development and constitute a distinct set of biomarkers that characterize tumor subgroups (69). For instance, pediatric EPNs exhibit a CpG island methylator phenotype (CIMP). Such DNA hypermethylation is mediated by the polycomb group of proteins and targets differentiation genes that are transcriptionally controlled by H3K27 marks. Pharmacological compounds against these alterations proved to be efficacious in vitro and in vivo (61). Finally, vascular endothelial growth factor (VEGF) is an alternative noninvasive biomarker in children with brain tumors that reflects enhanced tumor angiogenesis (70). Sobol-Milejska et al. reported significantly elevated VEGF expression in blood samples of 106 children diagnosed with brain tumors in comparison to the control group. VEGF expression has also been found upregulated in UW402 MB cells upon hypoxia. In accordance, strong expression of VEGFR has been correlated with gadolinium enhancement in MRI of pediatric patients with MB, a feature with prognostic significance (70).
Molecular therapeutic targeting of pediatric brain tumors
Recent studies on pediatric brain tumors treatment based on their specific genetic background have revealed several efficient molecular targets (Table 1). For example, in tuberous sclerosis-associated subependymal giant cell tumors, the mTOR pathway inhibitor everolimus has been tested with beneficial treatment outcome (79) and is currently further evaluated in LGGs (80). Other molecular targets including BRAF V600E and MEK inhibitors such as vemurafenib, dabrafenib and trametinib may enable the management of brain tumors. These inhibitors appear to have improved brain penetration and they could result in shrinkage of brain tumors (80). However, the study of Sievert et al. showed that BRAF inhibitors like Dabrafenib and PLX4720, can cause paradoxical stimulation of MAPK signaling in other BRAF mutations, like the fusion BRAF/KIAA1549 gene (81). Selumetinib, an efficient inhibitor of MAP kinase pathway (MEK inhibitors) could be an additional option for patients with LGG (82).
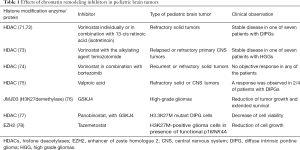
Full table
Recent studies have demonstrated that mTOR pathway upregulation may be related with malignant pLGGs. Rapalog everolimus could be used for mTOR-driven tumors, since it has successfully treated SEGAs in children with tuberous sclerosis (83). Furthermore, several novel targeted approaches such as, BRAF V600E, Ras/Akt pathway and telomerase are under investigation (84). In MYC-driven MB models, JQ, which is a BET BRD inhibitor was shown to be efficient (53) and in SMO inhibitor-resistant SHH MB with potential efficiency in GFI1/1B-activated MBs (85,86). Another SMO inhibitor tested specifically in patients with MB is the vismodegib, as well as sonidegib (87).
High-throughput screening has revealed the potential effectiveness of pemetrexed and gemcitabine and HDAC/PI3K inhibitors for group 3 MBs with the worst prognosis, being currently under clinical trial and expected that PF-EPN-A and histone-mutant HGGs will be responsive to epigenetic regulators (88). To date, the only clinically validated therapies that have emerged from molecular analysis of pediatric brain neoplasms are SMO antagonists, which show activity in patients with SHH-MB resulting from mutations in PTCH or SMO. The efficacy of these approaches remains to be validated in patients, but this strategy—using animal models (including GEMMs and PDXs) to detect and examine therapies for distinct types of brain tumors—holds great promise. Genetic characterization of PF EPN and MB (groups 3 and 4) is highly demanded in order to improve patients’ prognosis.
In another study, Taylor et al. reported that heterozygous somatic mutations in ACVR1 gene, which encodes the activin A type I receptor serine/threonine kinase ALK2 were found in 21% of pediatric DIPG (26). ACVR1 mutations were found to co-segregate with histone H3.1 K27M mutated DIPG (26). Previously, in patients with the autosomal dominant congenital childhood developmental disorder fibrodysplasia ossificans progressive (FOP), identical ACVR1 mutations have been suggested to constitutively activate the bone morphogenic protein (BMP)-dependent transforming growth factor-β pathway (89). This study proposes a role for BMP inhibitors to target one of the potential tumorigenic mechanisms in DIPG (26). Future trials would be of interest to see the efficacy of single BMP inhibitors or combined with epigenetic targeted therapies such as HDAC or EZH2 inhibitors.
Epigenetic targeting of histone modifications in HGGs using HDAC inhibitors
In many cancer types, HDACs are overexpressed. Therefore, using inhibitors to target them may contribute to the development of novel therapeutic schemes for pediatric brain tumors (Table 1). A phase I clinical trial performed by the Children’s Oncology Group, investigated the pan-HDAC inhibitor vorinostat individually or in combination with isotretinoin in children with refractory solid tumors (71).
The clinical observations detected prolonged stable disease in one of seven patients with DIPGs (72). In the same trial, vorinostat was given in relapsed or refractory primary brain tumors along with the alkylating agent, temozolomide and 1/7 patients with HGGs exhibited stable disease (73). Another phase I trial examined vorinostat in combination with bortezomib which is a ubiquitin-proteasome pathway blocker, in children with recurrent or refractory solid tumors. The results showed no objective response in any of the patients (74). In addition, the HDAC inhibitor valproic acid was explored in children with refractory solid or CNS tumors and a response was observed in 2/4 of patients with DIPGs (one partial and one minor) (75). Ongoing clinical trials evaluate event-free survival in children with newly diagnosed HGGs and brainstem gliomas after treatment with valproic acid and radiotherapy, followed by bevacizumab. An ongoing phase II/III trial in children with HGG, investigates the event-free survival using vorinostat, or temozolomide, or bevacizumab in combination with radiotherapy, followed by treatment with bevacizumab and temozolomide (28).
Epigenetic targeting of histone modifications in HGGs using histone demethylase inhibition
The constant knowledge of epigenetic changes underlying HGGs could be targeted and reversed leading to therapy of these tumors (76). Apart of HDAC inhibition Hashizume et al. investigated a therapeutic approach of histone demethylase inhibition using the H3K27 demethylase, JMJD3, with GSKJ4 in pHGGs (76). The increase of H3K27 methylation, is leading to gliomagenesis by inhibiting gene expression and blocking cell differentiation (90). In H3.3 K27M glioma cell lines, GSKJ4 treatment displayed a 50% reduction in growth, increased apoptosis, and inhibition of clonogenicity, while JMJD3-depleted glioma cells exhibited no significant decrease in proliferation (76). In athymic brainstem K27M glioma murine xenografts (nu/nu, BALB/c), GSKJ4 treatment reduced significantly tumor growth and prolonged survival (76). The pan-HDAC inhibitor panobinostat, was used with the histone demethylase inhibitor GSKJ4 in 14 patient-derived DIPG cell cultures (77). Cells expressing the H3.3 K27M mutation exhibited elevated H3 acetylation and H3K27 methylation after treatment, indicating a partial rescue of the H3.3 K27M-induced global hypomethylator phenotype CHOP. Additionally, in H3.3 K27M mutant DIPG cells, panobinostat was shown to act synergistically with GSKJ4 to decrease their viability (77), indicating histone methylation and acetylation targeting as an exciting treatment option for HGGs.
EZH2 inhibition is also an alternative mechanism to prevent abnormal histone methylation of target genes and could promote cell differentiation, while reducing cell proliferation in different tissues (91). Preclinical studies have tested pharmacological inhibition of EZH2 in pediatric rhabdomyosarcoma (92). EZH2 overexpression is reported in many malignancies including breast cancer, lymphoma, and prostate cancer (93,94). Clinical trials are currently investigating EZH2 inhibitors for the treatment of children with MB and EPN which display elevated PRC2 activity (56,61,95). Finally, tazemetostat, an EZH2 inhibitor, was shown to affect H3K27M-positive glioma cell growth in the presence of functional p16INK4A (78).
Conclusions
Experimental data over the last decade have changed entirely our view on the genomic and epigenetic landscape of pediatric brain neoplasms. Molecular and epigenetic biomarkers play a crucial role in tumor classification and novel therapeutic approaches. The identification of histone modifications that are correlated with the development of brain tumors in children has stimulated the investigation of epigenetic inhibitors such as histone demethylase and HDAC inhibitors in cancer treatment. However, the complex interaction between DNA methylation, histone modifications, gene expression, and chromatin organization presents a challenge for the design of rational trials (28). For instance, there are some restrictions in epigenetic targeting including elucidation of the underlying molecular mechanism or the effects of HDAC inhibitors on cellular signaling and pathways which are still unclear. Moreover, HGGs may be protected from damage due to intratumoral genetic heterogeneity, which could change the intracellular concentration and recruitment of the HDAC inhibitor. Furthermore, another limitation is the non-effective penetration of the blood-brain barrier of HDAC inhibitors such as vorinostat and panobinostat, hence they are failing to translate into efficient treatment (28,96). These reasons possibly explain the slow advance of clinical trials investigating HDAC inhibitors use in HGGs. The most important goal for the future will be to apply the molecular information of tumor’s subgroups in the identification of subgroup-specific or patient-specific therapies. In the long run, it is likely to use genomic, transcriptomic, and epigenetic information not only to predict patient outcomes but also to guide selection of the most efficient with the least toxicity forms of therapy (28).
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Chamdine O, Gajjar A. Molecular characteristics of pediatric high-grade gliomas. CNS Oncol 2014;3:433-43. [Crossref] [PubMed]
- Kramm CM, Butenhoff S, Rausche U, et al. Thalamic high-grade gliomas in children: a distinct clinical subset? Neuro Oncol 2011;13:680-9. [Crossref] [PubMed]
- Paugh BS, Qu C, Jones C, et al. Integrated Molecular Genetic Profiling of Pediatric High-Grade Gliomas Reveals Key Differences With the Adult Disease. J Clin Oncol 2010;28:3061-8. [Crossref] [PubMed]
- Forst DA, Nahed B V, Loeffler JS, et al. Low-grade gliomas. Oncologist 2014;19:403-13. [Crossref] [PubMed]
- Penman CL, Faulkner C, Lowis SP, et al. Current Understanding of BRAF Alterations in Diagnosis, Prognosis, and Therapeutic Targeting in Pediatric Low-Grade Gliomas. Front Oncol 2015;5:54. [Crossref] [PubMed]
- Johnson KJ, Cullen J, Barnholtz-Sloan JS, et al. Childhood brain tumor epidemiology: a brain tumor epidemiology consortium review. Cancer Epidemiol Biomarkers Prev 2014;23:2716-36. [Crossref] [PubMed]
- Dang M, Phillips PC. Pediatric Brain Tumors. Continuum (Minneap Minn) 2017;23:1727-57. [Crossref] [PubMed]
- Maury E, Hashizume R. Epigenetic modification in chromatin machinery and its deregulation in pediatric brain tumors: Insight into epigenetic therapies. Epigenetics 2017;12:353-69. [Crossref] [PubMed]
- Delcuve GP, Rastegar M, Davie JR. Epigenetic control. J Cell Physiol 2009;219:243-50. [Crossref] [PubMed]
- Faria CMC, Rutka JT, Smith C, et al. Epigenetic mechanisms regulating neural development and pediatric brain tumor formation. J Neurosurg Pediatr 2011;8:119-32. [Crossref] [PubMed]
- Haider BA, Baras AS, McCall MN, et al. A critical evaluation of microRNA biomarkers in non-neoplastic disease. PLoS One 2014;9. [Crossref] [PubMed]
- Bergthold G, Bandopadhayay P, Bi WL, et al. Pediatric low-grade gliomas: how modern biology reshapes the clinical field. Biochim Biophys Acta 2014;1845:294-307. [PubMed]
- Olow A, Mueller S, Yang X, et al. BRAF Status in Personalizing Treatment Approaches for Pediatric Gliomas. Clin Cancer Res 2016;22:5312-21. [Crossref] [PubMed]
- Klonou A, Piperi C, Gargalionis AN, et al. Molecular Basis of Pediatric Brain Tumors. Neuromolecular Med 2017;19:256-70. [Crossref] [PubMed]
- Raabe E, Kieran MW, Cohen KJ. New Strategies in Pediatric Gliomas: Molecular Advances in Pediatric Low-Grade Gliomas as a Model. Clin Cancer Res 2013;19:4553-8. [Crossref] [PubMed]
- Pópulo H, Lopes JM, Soares P. The mTOR Signalling Pathway in Human Cancer. Int J Mol Sci 2012;13:1886-918. [Crossref] [PubMed]
- Johnson A, Severson E, Gay L, et al. Comprehensive Genomic Profiling of 282 Pediatric Low- and High- Grade Gliomas Reveals Genomic Drivers, Tumor Mutational Burden, and Hypermutation Signatures. Oncologist 2017;22:1478-90. [Crossref] [PubMed]
- Le LQ, Parada LF. Tumor microenvironment and neurofibromatosis type I: connecting the GAPs. Oncogene 2007;26:4609-16. [Crossref] [PubMed]
- Sturm D, Witt H, Hovestadt V, et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell 2012;22:425-37. [Crossref] [PubMed]
- Rizzo D, Ruggiero A, Martini M, et al. Molecular Biology in Pediatric High-Grade Glioma: Impact on Prognosis and Treatment. Biomed Res Int 2015;2015. [Crossref] [PubMed]
- Schwartzentruber J, Korshunov A, Liu X-Y, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012;482:226-31. [Crossref] [PubMed]
- Wu G, Broniscer A, McEachron TA, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet 2012;44:251-3. [Crossref] [PubMed]
- Mackay A, Burford A, Carvalho D, et al. Integrated Molecular Meta-Analysis of 1,000 Pediatric High-Grade and Diffuse Intrinsic Pontine Glioma. Cancer Cell 2017;32:520-37.e5. [Crossref] [PubMed]
- Khuong-Quang DA, Buczkowicz P, Rakopoulos P, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol 2012;124:439-47. [Crossref] [PubMed]
- Fontebasso AM, Papillon-Cavanagh S, Schwartzentruber J, et al. Recurrent somatic mutations in ACVR1 in pediatric midline high-grade astrocytoma. Nat Genet 2014;46:462-6. [Crossref] [PubMed]
- Taylor KR, Mackay A, Truffaux N, et al. Recurrent activating ACVR1 mutations in diffuse intrinsic pontine glioma. Nat Genet 2014;46:457-61. [Crossref] [PubMed]
- Jones C, Perryman L, Hargrave D. Paediatric and adult malignant glioma: close relatives or distant cousins? Nat Rev Clin Oncol 2012;9:400-13. [Crossref] [PubMed]
- Williams MJ, Singleton WGB, Lowis SP, et al. Therapeutic Targeting of Histone Modifications in Adult and Pediatric High-Grade Glioma. Front Oncol 2017;7:45. [Crossref] [PubMed]
- Fontebasso AM, Schwartzentruber J, Khuong-Quang D-A, et al. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol 2013;125:659-69. [Crossref] [PubMed]
- Panditharatna E, Yaeger K, Kilburn LB, et al. Clinicopathology of diffuse intrinsic pontine glioma and its redefined genomic and epigenomic landscape. Cancer Genetics 2015;208:367-73. [Crossref] [PubMed]
- Bjerke L, Mackay A, Nandhabalan M, et al. Histone H3.3. mutations drive pediatric glioblastoma through upregulation of MYCN. Cancer Discov 2013;3:512-9. [Crossref] [PubMed]
- Saratsis AM, Kambhampati M, Snyder K, et al. Comparative multidimensional molecular analyses of pediatric diffuse intrinsic pontine glioma reveals distinct molecular subtypes. Acta Neuropathol 2014;127:881-95. [Crossref] [PubMed]
- Swartling FJ, Savov V, Persson AI, et al. Distinct neural stem cell populations give rise to disparate brain tumors in response to N-MYC. Cancer Cell 2012;21:601-13. [Crossref] [PubMed]
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer 2004;4:143-53. [Crossref] [PubMed]
- Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006;125:315-26. [Crossref] [PubMed]
- Ku M, Koche RP, Rheinbay E, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 2008;4. [Crossref] [PubMed]
- Lewis PW, Müller MM, Koletsky MS, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 2013;340:857-61. [Crossref] [PubMed]
- Venneti S, Garimella MT, Sullivan LM, et al. Evaluation of histone 3 lysine 27 trimethylation (H3K27me3) and enhancer of Zest 2 (EZH2) in pediatric glial and glioneuronal tumors shows decreased H3K27me3 in H3F3A K27M mutant glioblastomas. Brain Pathol 2013;23:558-64. [Crossref] [PubMed]
- Bender S, Tang Y, Lindroth AM, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer Cell 2013;24:660-72. [Crossref] [PubMed]
- Chan KM, Fang D, Gan H, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev 2013;27:985-90. [Crossref] [PubMed]
- Schwalbe EC, Lindsey JC, Nakjang S, et al. Novel molecular subgroups for clinical classification and outcome prediction in childhood medulloblastoma: a cohort study. Lancet Oncol 2017;18:958-71. [Crossref] [PubMed]
- Kool M, Koster J, Bunt J, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. Hide W, editor. PLoS One 2008;3:e3088.
- DeSouza RM, Jones BRT, Lowis SP, et al. Pediatric medulloblastoma - update on molecular classification driving targeted therapies. Front Oncol 2014;4:176. [Crossref] [PubMed]
- Northcott PA, Buchhalter I, Morrissy AS, et al. The whole-genome landscape of medulloblastoma subtypes. Nature 2017;547:311-7. [Crossref] [PubMed]
- Kool M, Korshunov A, Remke M, et al. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol 2012;123:473-84. [Crossref] [PubMed]
- Taylor MD, Northcott PA, Korshunov A, et al. Molecular subgroups of medulloblastoma: the current consensus. Acta Neuropathol 2012;123:465-72. [Crossref] [PubMed]
- Liu KW, Pajtler KW, Worst BC, et al. Molecular mechanisms and therapeutic targets in pediatric brain tumors. Sci Signal 2017;10. [Crossref] [PubMed]
- Roussel MF, Stripay JL. Epigenetic Drivers in Pediatric Medulloblastoma. Cerebellum 2018;17:28-36. [Crossref] [PubMed]
- Northcott PA, Nakahara Y, Wu X, et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat Genet 2009;41:465-72. [Crossref] [PubMed]
- Pfister S, Rea S, Taipale M, et al. The histone acetyltransferase hMOF is frequently downregulated in primary breast carcinoma and medulloblastoma and constitutes a biomarker for clinical outcome in medulloblastoma. Int J cancer 2008;122:1207-13. [Crossref] [PubMed]
- Milde T, Oehme I, Korshunov A, et al. HDAC5 and HDAC9 in medulloblastoma: novel markers for risk stratification and role in tumor cell growth. Clin Cancer Res 2010;16:3240-52. [Crossref] [PubMed]
- Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell 2014;54:728-36. [Crossref] [PubMed]
- Bandopadhayay P, Bergthold G, Nguyen B, et al. BET bromodomain inhibition of MYC-amplified medulloblastoma. Clin Cancer Res 2014;20:912-25. [Crossref] [PubMed]
- Jones DTW, Northcott PA, Kool M, et al. The role of chromatin remodeling in medulloblastoma. Brain Pathol 2013;23:193-9. [Crossref] [PubMed]
- Schuettengruber B, Martinez AM, Iovino N, et al. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol 2011;12:799-814. [Crossref] [PubMed]
- Dubuc AM, Remke M, Korshunov A, et al. Aberrant patterns of H3K4 and H3K27 histone lysine methylation occur across subgroups in medulloblastoma. Acta Neuropathol 2013;125:373-84. [Crossref] [PubMed]
- Bunt J, Hasselt NA, Zwijnenburg DA, et al. OTX2 sustains a bivalent-like state of OTX2-bound promoters in medulloblastoma by maintaining their H3K27me3 levels. Acta Neuropathol 2013;125:385-94. [Crossref] [PubMed]
- Bunt J, Hasselt NE, Zwijnenburg DA, et al. OTX2 directly activates cell cycle genes and inhibits differentiation in medulloblastoma cells. Int J cancer 2012;131:E21-32. [Crossref] [PubMed]
- Parker M, Mohankumar KM, Punchihewa C, et al. C11orf95-RELA fusions drive oncogenic NF-κB signalling in ependymoma. Nature 2014;506:451-5. [Crossref] [PubMed]
- Bayliss J, Mukherjee P, Lu C, et al. Lowered H3K27me3 and DNA hypomethylation define poorly prognostic pediatric posterior fossa ependymomas. Sci Transl Med 2016;8. [Crossref] [PubMed]
- Mack SC, Witt H, Piro RM, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature 2014;506:445-50. [Crossref] [PubMed]
- Reddington JP, Perricone SM, Nestor CE, et al. Redistribution of H3K27me3 upon DNA hypomethylation results in de-repression of Polycomb target genes. Genome Biol 2013;14:R25. [Crossref] [PubMed]
- Mendenhall EM, Koche RP, Truong T, et al. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet 2010;6. [Crossref] [PubMed]
- Huse JT, Rosenblum MK. The Emerging Molecular Foundations of Pediatric Brain Tumors. J Child Neurol 2015;30:1838-50. [Crossref] [PubMed]
- Staedtke V, Dzaye OD, Holdhoff M. Actionable Molecular Biomarkers in Primary Brain Tumors. Trends Cancer 2016;2:338-49. [Crossref]
- Liu XY, Gerges N, Korshunov A, et al. Frequent ATRX mutations and loss of expression in adult diffuse astrocytic tumors carrying IDH1/IDH2 and TP53 mutations. Acta Neuropathol 2012;124:615-25. [Crossref] [PubMed]
- Paugh BS, Zhu X, Qu C, et al. Novel oncogenic PDGFRA mutations in pediatric high-grade gliomas. Cancer Res 2013;73:6219-29. [Crossref] [PubMed]
- Pérez-Ramírez M, Hernández-Jiménez AJ, Guerrero-Guerrero A, et al. Genomics and epigenetics: A study of ependymomas in pediatric patients. Clin Neurol Neurosurg 2016;144:53-8. [Crossref] [PubMed]
- Hovestadt V, Jones DTW, Picelli S, et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature 2014;510:537-41. [Crossref] [PubMed]
- Sobol-Milejska G, Mizia-Malarz A, Musiol K, et al. Serum levels of vascular endothelial growth factor and basic fibroblast growth factor in children with brain tumors. Adv Clin Exp Med 2017;26:571-5. [Crossref] [PubMed]
- Fouladi M, Park JR, Stewart CF, et al. Pediatric phase I trial and pharmacokinetic study of vorinostat: a Children’s Oncology Group phase I consortium report. J Clin Oncol 2010;28:3623-9. [Crossref] [PubMed]
- Hummel TR, Wagner L, Ahern C, et al. A pediatric phase 1 trial of vorinostat and temozolomide in relapsed or refractory primary brain or spinal cord tumors: A children’s oncology group phase 1 consortium study. Pediatr Blood Cancer 2013;60:1452-7. [Crossref] [PubMed]
- Bezecny P. Histone deacetylase inhibitors in glioblastoma: pre-clinical and clinical experience. Med Oncol 2014;31:985. [Crossref] [PubMed]
- Muscal JA, Thompson PA, Horton TM, et al. A phase I trial of vorinostat and bortezomib in children with refractory or recurrent solid tumors: a Children’s Oncology Group phase I consortium study (ADVL0916). Pediatr Blood Cancer 2013;60:390-5. [Crossref] [PubMed]
- Su JM, Li XN, Thompson P, et al. Phase 1 study of valproic acid in pediatric patients with refractory solid or CNS tumors: a children’s oncology group report. Clin Cancer Res 2011;17:589-97. [Crossref] [PubMed]
- Hashizume R, Andor N, Ihara Y, et al. Pharmacologic inhibition of histone demethylation as a therapy for pediatric brainstem glioma. Nat Med 2014;20:1394-6. [Crossref] [PubMed]
- Grasso CS, Tang Y, Truffaux N, et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med 2015;21:827. [Crossref] [PubMed]
- Mohammad F, Weissmann S, Leblanc B, et al. EZH2 is a potential therapeutic target for H3K27M-mutant pediatric gliomas. Nat Med 2017;23:483-92. [Crossref] [PubMed]
- Krueger DA, Care MM, Agricola K, et al. Everolimus long-term safety and efficacy in subependymal giant cell astrocytoma. Neurology 2013;80:574-80. [Crossref] [PubMed]
- Gajjar A, Bowers DC, Karajannis MA, et al. Pediatric Brain Tumors: Innovative Genomic Information Is Transforming the Diagnostic and Clinical Landscape. J Clin Oncol 2015;33:2986-98. [Crossref] [PubMed]
- Sievert AJ, Lang SS, Boucher KL, et al. Paradoxical activation and RAF inhibitor resistance of BRAF protein kinase fusions characterizing pediatric astrocytomas. Proc Natl Acad Sci U S A 2013;110:5957-62. [Crossref] [PubMed]
- Kolb EA, Gorlick R, Houghton PJ, et al. Initial testing (stage 1) of AZD6244 (ARRY-142886) by the Pediatric Preclinical Testing Program. Pediatr Blood Cancer 2010;55:668-77. [Crossref] [PubMed]
- Garcia MA, Solomon DA, Haas-Kogan DA. Exploiting molecular biology for diagnosis and targeted management of pediatric low-grade gliomas. Future Oncol 2016;12:1493-506. [Crossref] [PubMed]
- Wells EM, Packer RJ. Pediatric brain tumors. Continuum (Minneap Minn) 2015;21:373-96. [Crossref] [PubMed]
- Northcott PA, Lee C, Zichner T, et al. Enhancer hijacking activates GFI1 family oncogenes in medulloblastoma. Nature 2014;511:428-34. [Crossref] [PubMed]
- Tang Y, Gholamin S, Schubert S, et al. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nat Med 2014;20:732-40. [Crossref] [PubMed]
- Kieran MW. Targeted treatment for sonic hedgehog-dependent medulloblastoma. Neuro Oncol 2014;16:1037-47. [Crossref] [PubMed]
- Wu G, Diaz AK, Paugh BS, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet 2014;46:444-50. [Crossref] [PubMed]
- Kaplan FS, Xu M, Seemann P, et al. Classic and atypical fibrodysplasia ossificans progressiva (FOP) phenotypes are caused by mutations in the bone morphogenetic protein (BMP) type I receptor ACVR1. Hum Mutat 2009;30:379-90. [Crossref] [PubMed]
- Morales La Madrid A, Hashizume R, Kieran MW. Future Clinical Trials in DIPG: Bringing Epigenetics to the Clinic. Front Oncol 2015;5:148. [Crossref] [PubMed]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer 2006;6:846-56. [Crossref] [PubMed]
- Ciarapica R, Carcarino E, Adesso L, et al. Pharmacological inhibition of EZH2 as a promising differentiation therapy in embryonal RMS. BMC Cancer 2014;14:139. [Crossref] [PubMed]
- Kleer CG, Cao Q, Varambally S, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A 2003;100:11606-11. [Crossref] [PubMed]
- Sneeringer CJ, Scott MP, Kuntz KW, et al. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci U S A 2010;107:20980-5. [Crossref] [PubMed]
- Robinson G, Parker M, Kranenburg TA, et al. Novel mutations target distinct subgroups of medulloblastoma. Nature 2012;488:43-8. [Crossref] [PubMed]
- Veringa SJ, Biesmans D, van Vuurden DG, et al. In vitro drug response and efflux transporters associated with drug resistance in pediatric high grade glioma and diffuse intrinsic pontine glioma. PLoS One 2013;8. [Crossref] [PubMed]


