Liquid biopsies in lung cancer—time to implement research technologies in routine care?
Lung cancer is the leading cause of cancer mortality causing 1.6 million deaths annually (1). During the last decade, a substantial progress in the understanding of lung cancer biology has resulted in several promising targeted therapies for advanced disease (2). Most lung cancer patients are diagnosed at a late stage where systemic medical treatment is the treatment of choice. The druggable targets discovered so far include point mutations in important genes such as EGFR, BRAF, HER2, PI3K and re-arrangements in genes such as ALK, ROS1 and MET (3,4).
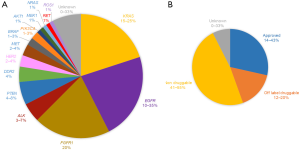
Molecular pathology using next generation sequencing (NGS) among other methods is now introduced in routine lung cancer care worldwide. Specific driver or candidate driver mutations are detected in more than half of lung cancer patients today, including point mutations in KRAS and EGFR, amplifications in FGFR1 and re-arrangements in ALK as the most common sequence variants (Figure 1A). Targeted therapy for patients having EGFR mutations or re-arrangements in ALK or ROS1 is approved for first line use in advanced disease (5) (Figure 1B). Insights in the mechanisms of resistance to targeted therapies in lung cancer have led to specific second line treatments for specific EGFR and ALK mutations (6,7). The need for re-biopsy procedures in lung cancer patients is thereby increasing, implicating a potential problem in lung cancer care since re-biopsies are linked to risk for morbidity and represents a logistical problem in many health care systems where resources for transthoracic biopsies are limited. The possibility to monitor lung cancer through blood based tests is therefore an important field where rapid technological developments make great promise for the future.
It is today possible to sequence the whole genome of a child from the mother’s blood (8) or find chromosomal abnormalities even in early cancer (9). Genomic analysis of tumor tissue through NGS is already being implemented in personalized medicine approaches for cancer patients, where oncogenic drivers are identified from tumor tissue. This have changed our way to think about genomic alterations and cancer, and ongoing studies such as the “Basket trial” (10) have identified and directly acted on targetable alterations to decide therapy rather than to treat patients accordingly to their cancer type (e.g., lung or breast cancer). This approach will further drive the liquid biopsy field, since most advanced cancers have already started to disseminate sub-clones/metastases far from the primary tumor and where one needle biopsy will likely not be representable for the full complexity of tumor heterogeneity (11). Liquid biopsies on the other hand reflect secreted information from the whole body, including tumor information, and are therefore not biased by analyses of only a small fraction of the tumor (a needle core). Liquid biopsies may however be limited by accessibility of tumor-derived material in less advanced cancers. Another limiting step will be the vast amount of information gathered and how to separate information derived from the tumor from information from normal healthy cells or adversely reacting non-tumorous tissue during cancer treatment.
This review focuses on future perspectives of liquid biopsies in lung cancer care for different clinical settings and present current technological platforms for further discussion of possible strategies for implementation of liquid biopsies in lung cancer.
Biosources
Whether the biosource is blood, sputum, urine, or saliva they all have different compositions of secreted material from the tumor, also dependent on disease type. Identifying a one-for-all biosource may not be possible, as it may be advantageous to combine biosources to neutralize potential issues and enhance the clinical possibilities. The amount of genetic information that can be generated from a single blood specimen is enormous and to cipher through the data requires computing power and skilled bioinformatics. One way of reducing the information is to isolate sub-fractions of a biosource to enrich for biomarkers of interest.
Circulating tumor cells (CTCs)
A full cancer genome and transcriptome is present in each individual CTC and protein markers can be used for isolation or enumeration of them, therefore they represent an interesting source of tumor information where mutations and gene fusions can be found (12-14). Many studies have used enumeration of CTCs to predict recurrence or used cancer-derived biomarkers to follow therapy resistance (12,15). However, elaborate techniques are required to isolate pure CTCs as residual immune cells often contaminate the CTC population. One obvious drawback is the relatively low abundance of CTCs in the circulation even in advanced disease.
Extracellular vesicles (EVs)
EVs are membrane vesicles derived from plasma membrane shedding or released through the multi-vesicular body compartments, their size varies from ~30 to several hundred nm. They have been shown to transfer bioactive molecules between cells and organs, with the potential to alter the immune system or facilitate metastatic niches formation (16,17). These very numerous entities carry information from the host cell, in terms of DNA, mRNA, miRNA or protein molecules. The most abundant EVs are platelet derived and isolation of pure tumor exosomes remains a big hurdle, since not only normal EVs but also protein- and lipid complexes may interfere with the isolation process (18,19).
Platelets
An often-neglected blood entity, platelets, has emerged as a novel biosource for tumor-derived material (20,21), and are known to sequester material during their circulation. Platelets have a relatively long time in circulation, 4 to 6 days, and continuously sequestering material throughout their passage. Their transcriptome also changes on environmental cues and the platelets thereby become educated by the current state of the individual. Using the education process, it has been possible to accurately predict whether cancer is present, the type of cancer, therapy response and oncogenic alterations driving the disease (21-23).
Cell-free circulating tumor DNA (ctDNA)/cell-free circulating DNA (cfDNA)
The interest for ctDNA has risen concomitant with the improvement of power of sequencing, and is now the dominating liquid biopsy platform in terms of utility and sensitivity. Where traditional Sanger sequencing could identify genomic mutations present in more than 5% of the alleles, it is now possible with adequate coverage and error elimination techniques to identify mutations at a sensitivity as good as digital PCR (dPCR) in broad gene panels (24). Apart from mutations, also structural alterations (25), methylation patterns (26) or nucleosomal footprints (27) may add additional information on tumor load and site of tumor origin. ctDNA releases into the circulation has been attributed to cellular decay mechanisms like apoptosis or necrosis, with a distinct pattern in blood corresponding to DNA fragmentation around nucleosome units (28). In advanced disease >10% of circulating DNA can be traced back to the tumor genome (29). However, in early disease very little ctDNA (picogram rather than nanogram amounts) is present in the circulation. This increases the demands of the technology, pushing it to the limit regarding sensitivity and specificity.
Technology
Technological advances in detection of low frequency mutations and lowering of costs have made it possible to reach sensitivities below 0.1%, with several platforms available for detection of low frequency mutation variants in ctDNA (30). These include real-time quantitative PCR (qPCR), NGS, dPCR, and beads-emulsion-amplification-and-magnetics (BEAMing). ctDNA is the biosource commonly used in clinical routine, although the mentioned techniques have application potential also to CTCs, EVs and platelets. Detection of CTCs relies on physical (size) or biological properties (cell surface markers) and the CTC count can be used as a diagnostic and prognostic marker. The technological focus in this review is on analysis of ctDNA with brief summaries of available methods below.
qPCR
Screening for variants in ctDNA using qPCR is fast and relatively inexpensive. qPCR is commonly performed with sequence-specific probes that are labelled with a fluorescent reporter which allows detection only after hybridization of the probe with its complementary sequence. The amount of amplified product is linked to fluorescence intensity. It is a straightforward test, however with a sensitivity of about 10% (30) it is unable to match the high sensitivity of other techniques such as dPCR or NGS.
dPCR
The dPCR approach follows the same reaction principle as qPCR, although here the samples are partitioned into thousands of parallel PCR reactions. The systems can either be array or droplet based. The array system BioMark by Fluidigm partitions the DNA templates to chambers were the PCR reactions can be monitored in real time. The droplet-based platforms ddPCRTM by Bio-Rad and RainDropTM by RainDance Technologies dispenses the PCR reaction mix and DNA templates into 20,000 and 10,000,000 droplets, respectively, and the presence or absence of fluorescence in each partition is then used to calculate the absolute number of targets present in the original sample. The principle by which dPCR is performed where a single or a few templates are present in each chamber/droplet allows for detection of mutated ctDNA on a high background of wild type cfDNA and quantification of even small fold change differences is possible (31) yielding a sensitivity of <0.1%.
BEAMing
BEAMing is a combination of emulsion dPCR with magnetic beads and flow cytometry for detection. Sequence specific primers are used to amplify the targets and the amplicons become coupled to magnetic streptavidin beads before emulsion into droplets. The PCR fragments are subsequently released from the beads, and as with conventional dPCR, flow cytometry is then used to sort beads containing the mutation (32). The sensitivity of BEAMing is analogous to dPCR (33) although comes with a more complex protocol. Furthermore, qPCR, dPCR, and BEAMing have in common that they can only screen for known mutations.
NGS
NGS involves massively parallel sequencing where millions or billions of DNA templates are sequenced simultaneously. The major advantage of NGS over the previously described methods is that it is possible to screen for unknown sequence variants. With whole-genome-sequencing (WGS) it is possible to get the full genetic profile of a tumor, including point mutations, small insertions/deletions, rearrangements, and copy number aberrations from nanograms of DNA. Whole-exome-sequencing (WES) provides information of all coding bases of the genome typically for a lesser price per base than WGS. WES is however still almost out of grasp for ctDNA due to that sample input requirements is usually hundreds of nanograms. Targeted sequencing, on the other hand, with gene panels consisting of tens to hundreds of genes is an appealing approach for ctDNA analysis. The cost is manageable for clinical applications and gene panels covering the major genes for the specific tumor type can be constructed from the scientific literature.
The sensitivity of NGS is <1% (34,35) but efforts have been made to decrease this number as well as increase the specificity using unique molecular identifiers (UMIs). These are random stretches of nucleotides that are incorporated in the DNA fragments to be sequenced before amplification. The UMI acts as a molecular memory of the number of molecules in the starting sample so that these can be accurately quantified after amplification and sequencing (36). Furthermore, a true variant will be present in the majority of PCR progenies with the same UMI whereas variants that are artefacts will not be shared by the other PCR progenies from the same founder molecule. Hence, a consensus call can be made after grouping PCR progenies of the same original molecule. In all, this results in that variants can be called accurately at lower frequencies compared to a library that is constructed without UMIs.
NGS is still an expensive technique and is also time-consuming. Therefore, an NGS approach is suitable at the time of diagnosis to get a detailed view of the genetic architecture of the cancer, while at follow-up a specific and sensitive, but less expensive and quicker methods such as dPCR may be a good choice.
Nanostring
The nCounter® technology (NanoString® Technologies) is emerging as a method to screen for the clinically relevant ALK, ROS1, and RET fusion genes in lung cancer tissue samples (37). The nCounter technology employs a strategy where two probes bind to the target RNA molecule. One of the probes contains a fluorescent reporter and the second a biotinylated capture probe allowing for immobilization to the cartridge surface. During imaging the color code of the reporters are counted for each target molecule. This is a method that holds promises also for circulating RNA. While until now it has mainly been used to detect quantitative differences in levels of miRNA from plasma, it has been shown that sufficient amounts of RNA can be extracted from plasma and used for screening by this technique (38,39).
Discussion
More sensitive assays for biomarker identification also require increased knowledge of the biosource that is explored. Like the rapid progression of ultra-deep sequencing of samples where normal individuals may display genetic aberrations of minimal frequency or silent modifications that reside in so far healthy cells or representing very early changes (40-42). Measuring circulatory tumor burden will be vital to identify and follow tumor growth, especially in early cancer were tumor type and location may elude the clinicians, but the genetic alterations found in ctDNA can be tracked in real-time discriminating between fast-growing life-threatening tumors from more indolent slow growing tumors or premalignant growth that are of no concern for the wellbeing of the patient. Liquid biopsies are quickly moving from research into clinical practice for advanced disease, especially in NSCLC that has many genetic alterations that provide therapeutic opportunities (Figure 1B). We can all foresee a day when liquid biopsies are used in daily routine in pathological evaluation situations, but one issue that has arisen is the sometimes poor concordance between analyses of a “real” tissue biopsy and the “liquid” biopsy (43,44). The studies highlighted a heterogeneity issue between matched tissue and blood based mutational profiles. What consequences that will have on decision making, which mutations are clinically actionable, and whether it is a result of tumor clonal heterogeneity or technical variation from usage of different sequencing platforms and bioinformatic workflows (11,45) remains to be seen.
In the discussion we focus on liquid biopsies in lung cancer care and clinical implementations of ctDNA detection through a cancer stage perspective.
Screening
Early diagnosis is usually beneficial to the lung cancer patients as it increases the possibility of curative treatment as surgery or stereotactic body radiotherapy (SBRT). In the large national lung cancer screening trial (NLST) a reduction of lung cancer mortality of 20% was observed in the experimental arm where low-dose CT screening of high risk populations where offered (46). Screening for lung cancer is today reimbursed by major insurance companies in the USA and Europe is on the rim of introducing screening programs. The lesson learned from the NLST trial is that the number of false positive nodules is very high and the need for invasive diagnostic procedures is high. Lung cancer screening is therefore aiming towards high risk populations selected on simple clinical factors such as age and smoking status. Blood based biomarkers defining populations with higher risk of developing lung cancer is therefore needed and analysis of circulating DNA or RNA may represent one possible way to reduce screening cohorts and increase the positive predictive value of screening.
Previous studies have shown the possibility to identify ctDNA and point mutations in healthy undiagnosed persons up to 5 years prior their cancer diagnosis (42), but also healthy persons that did not develop cancer had acquired mutations. This indicates that early cancer detection is possible, but extreme care needs to be taken to understand what these alterations mean and how they change overtime. Early changes may not be answered directly since no imaging modalities or alternative conformational sources are available to verify tumor growth, changes will earmark the patient for active monitoring with frequent follow ups to identify active tumor growth. Therefore, finding “a needle” in the haystack is not good enough, there is a great need to know what it represents. To this end different consortia and institutions have joined forces to build large data bases of the genomic distribution in liquid biopsies from cancer patients and the healthy population like the Blood Profiling Atlas in Cancer (47), part of the U.S. Cancer Moonshot initiative, and the Circulating Cell-free Genomic Atlas Study (CCGA) (48), to understand limitations and possibilities with blood based liquid biopsies.
Stage I–II
Early stage lung cancer is usually managed by surgery. Lobectomy or pneumonectomy is the gold standard of curative treatment for stage I–II patients. Patients inoperable due to concurrent disease may be offered SBRT with curative intention. However, the risk of relapse after surgery is still high and a majority of patients are also offered postoperative adjuvant chemotherapy. The impact of adjuvant chemotherapy is modest with an increase of 5-year survival of only 5% after four cycles of platinum-based chemotherapy (49). The number needed to treat to have an impact with adjuvant chemotherapy is therefore high and new prognostic biomarkers are needed for personalized treatment decisions in the adjuvant setting. Despite genetic profiling of tumor tissue, liquid biopsies will most certainly play an important role for this in the future, since there is a rather large possibility that no tumor material will be available due to biopsy issues, and repeated biopsies will increase the risk of major complications (50). Therefore, the possibility to use liquid biopsies as a complement to tissue biopsies and in the longitudinal follow up of early stage lung cancer is intriguing. Liquid biopsies may be of value in detecting minimal residual disease (MRD) (51), and adjustments of adjuvant treatment. Follow up after treatment of early stage lung cancer is today mainly based on radiology, but several studies using released tumor material could identify genetic alterations several weeks prior to radiologically verified recurrence (23,25), indicating that blood based-tests are warranted and that detecting circulating tumor-specific genes will be of great value in the follow-up of lung cancer patients. Tumor specific alterations are today observed in more than 50% of lung cancer cases and these genes may serve as circulating genetic markers for early relapse and to define current tumor burden.
Stage III
Locally advanced lung cancer constitutes approximately 25% of lung cancer cases worldwide. There is today no international gold standard in the treatment for these patients. The treatment options contain surgery with neo-adjuvant or adjuvant chemo or radiochemotherapy, or radiochemotherapy alone. Patients with widespread mediastinal metastasis unsuitable for radiochemotherapy or surgery are usually offered palliative treatment as for patients with advanced disease. The most common treatment approach with curative intention today is concomitant radiochemotherapy with a 5-year survival of approx. 15% (52). The toxicity of radiochemotherapy is significant with severe esophagitis and pneumonitis as the most frequent severe side-effects. Genetic profiling of lung cancer prior to curative radiochemotherapy may offer better prognostic and predictive factors for future stratification of patients where curative treatment is an option. Liquid biopsies may offer a possibility to continuously monitor tumor response during and after a course of radiochemotherapy. Quantification of circulating tumor specific genes may be a way to monitor tumor response early during the treatment and thus offer a theoretical possibility to adjust fractionation and dose after genetic response as assessed by liquid biopsies. Again, tumor specific genes detected up-front using NGS may be surrogate markers for tumor burden and treatment response. Liquid biopsies may also be of value in the follow up of patients treated with radiochemotherapy where identification of recurrence is mainly based on radiology and false positive findings due to pneumonitis or fibrosis is a clinical everyday problem. Today, targeted therapies are not commonly used in the treatment of locally advanced lung cancer, but ongoing clinical trials may re-introduce targeted therapies in the neo-adjuvant or adjuvant setting for patients with targetable genetic aberrations. In this future context liquid biopsies may be an important tool to monitor treatment as well as detect relapses.
Advanced disease
The majority of lung cancer patients are diagnosed with advanced disease where no curative treatments yet exist. The mainstay of treatment is still chemotherapy but a growing cohort is patients with specific druggable driver mutations such as EGFR, ALK, ROS1 and BRAF. Immunotherapy using antibodies inhibiting important checkpoints in lymphocyte activation is a new treatment modality implemented worldwide today where high mutational burden predicts a good therapy response (53). Genetic profiling of patients with advanced lung cancer is standard of care today. Assessment of EGFR mutations and ALK or ROS1 re-arrangements are mandatory according to most regulatory authorities (54). A number of genetic aberrations in genes such as BRAF, PI3K and HER2 are druggable using off-label treatments not yet approved for lung cancer (5) (Figure 1B). The value of targeted therapy in these patients is promising (5) but still to be confirmed in randomized trials. A proportion of patients have unknown driver mutations (Figure 1B). The increasing number of targetable genes in lung cancer is the main reason why PCR-based methods now are changed to different NGS based methods for genetic profiling of lung cancer up front. Besides being an alternative to invasive biopsies in selected cases, liquid biopsies offer a unique possibility to monitor treatment outcome during and after medical treatment. The most imminent impact of liquid biopsies is monitoring of targeted therapy where quantification of the target gene in circulation appears to correlate to treatment response (55). Resistance to commonly used targeted therapies is to a great extent caused by secondary point mutations emerging during treatment (56). For these patients repeated liquid biopsies offer a unique possibility to monitor early resistance development giving a possibility to modify the treatment after the actual genetic profile of the tumor (57). A number of second and third line EGFR and ALK TKIs with different activity against resistance mutations exist today making it possible to tailor treatment after the mutational spectrum. Liquid biopsies offer a less invasive method to monitor genetic changes as compared to repeated bronchoscopic or transthoracic biopsies (50). Quantification of circulating tumor specific genes may also be of predictive value as surrogate markers for patients during and after non-targeted treatment such as chemotherapy or immunotherapy.
Technical challenges
The rapid advance in the possibility to detect actionable specific genetic alternations of ctDNA in the diagnosis, treatment and follow-up of lung cancer has high-lighted the challenges of using liquid biopsies in routine clinical practice. A few studies have e.g., demonstrated the additional value of deep sequencing of ctDNA to detect driver and resistance mutations also when tissue was not available (58) whereas others studies have demonstrated a considerable lack of concordance between mutation analysis results of cancer tissue and liquid biopsies (43). The reasons for this discordance in output is not solely due to the biosource analyzed but may depend on the genetic test applied (44). In short, many issues have to be addressed in order to reach reliable clinical usability of liquid biopsies. These include clinical guidelines for sample collection and storage, cfDNA isolation, standardized quantification and genetic analysis as well as data processing, interpretation and validation. With time these issues will for sure be resolved and emphasize the advantages of liquid biopsies to detect actionable genetic alternations in the diagnosis and treatment of lung cancer.
Acknowledgements
Financial support was provided by Lion’s Cancer Research Foundation, Umeå University and the Swedish Research Council.
Footnote
Conflicts of Interest: JN is a shareholder of GRAIL. The other authors have no conflicts of interest to declare.
References
- Cheng TY, Cramb SM, Baade PD, et al. The international epidemiology of lung cancer: latest trends, disparities, and tumor characteristics. J Thorac Oncol 2016;11:1653-71. [Crossref] [PubMed]
- Hirsch FR, Scagliotti GV, Mulshine JL, et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299-311. [Crossref] [PubMed]
- Ortiz-Cuaran S, Scheffler M, Plenker D, et al. Heterogeneous mechanisms of primary and acquired resistance to third-generation EGFR inhibitors. Clin Cancer Res 2016;22:4837-47. [Crossref] [PubMed]
- Lovly C, Horn L, Pao W. Molecular Profiling of Lung Cancer. My Cancer Genome 2016. Available online: https://www.mycancergenome.org/content/disease/lung-cancer/
- Shea M, Costa DB, Rangachari D. Management of advanced non-small cell lung cancers with known mutations or rearrangements: latest evidence and treatment approaches. Ther Adv Respir Dis 2016;10:113-29. [Crossref] [PubMed]
- Konduri K, Gallant JN, Chae YK, et al. EGFR fusions as novel therapeutic targets in lung cancer. Cancer Discov 2016;6:601-11. [Crossref] [PubMed]
- Mok TS, Wu YL, Ahn MJ, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med 2017;376:629-40. [Crossref] [PubMed]
- Kitzman JO, Snyder MW, Ventura M, et al. Noninvasive whole-genome sequencing of a human fetus. Sci Transl Med 2012;4:137ra76. [Crossref] [PubMed]
- Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24. [Crossref] [PubMed]
- Lopez-Chavez A, Thomas A, Rajan A, et al. Molecular profiling and targeted therapy for advanced thoracic malignancies: a biomarker-derived, multiarm, multihistology phase II basket trial. J Clin Oncol 2015;33:1000-7. [Crossref] [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [Crossref] [PubMed]
- Antonarakis ES, Lu C, Wang H, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med 2014;371:1028-38. [Crossref] [PubMed]
- Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 2008;359:366-77. [Crossref] [PubMed]
- Park SM, Wong DJ, Ooi CC, et al. Molecular profiling of single circulating tumor cells from lung cancer patients. Proc Natl Acad Sci USA 2016;113:E8379-86. [Crossref] [PubMed]
- Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781-91. [Crossref] [PubMed]
- Hedlund M, Stenqvist AC, Nagaeva O, et al. Human placenta expresses and secretes NKG2D ligands via exosomes that down-modulate the cognate receptor expression: evidence for immunosuppressive function. J Immunol 2009;183:340-51. [Crossref] [PubMed]
- Peinado H, Aleckovic M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 2012;18:883-91. [Crossref] [PubMed]
- György B, Módos K, Pállinger E, et al. Detection and isolation of cell-derived microparticles are compromised by protein complexes resulting from shared biophysical parameters. Blood 2011;117:e39-48. [Crossref]
- Vickers KC, Palmisano BT, Shoucri BM, et al. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol 2011;13:423-33. [Crossref] [PubMed]
- Klement GL, Yip TT, Cassiola F, et al. Platelets actively sequester angiogenesis regulators. Blood 2009;113:2835-42. [Crossref] [PubMed]
- Nilsson RJ, Balaj L, Hulleman E, et al. Blood platelets contain tumor-derived RNA biomarkers. Blood 2011;118:3680-3. [Crossref] [PubMed]
- Best MG, Sol N, Kooi I, et al. RNA-Seq of tumor-educated platelets enables blood-based pan-cancer, multiclass, and molecular pathway cancer diagnostics. Cancer Cell 2015;28:666-76. [Crossref] [PubMed]
- Nilsson RJ, Karachaliou N, Berenguer J, et al. Rearranged EML4-ALK fusion transcripts sequester in circulating blood platelets and enable blood-based crizotinib response monitoring in non-small-cell lung cancer. Oncotarget 2016;7:1066-75. [Crossref] [PubMed]
- Kennedy SR, Schmitt MW, Fox EJ, et al. Detecting ultralow-frequency mutations by Duplex Sequencing. Nat Protoc 2014;9:2586-606. [Crossref] [PubMed]
- Reinert T, Scholer LV, Thomsen R, et al. Analysis of circulating tumour DNA to monitor disease burden following colorectal cancer surgery. Gut 2016;65:625-34. [Crossref] [PubMed]
- Lehmann-Werman R, Neiman D, Zemmour H, et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci USA 2016;113:E1826-34. [Crossref] [PubMed]
- Snyder MW, Kircher M, Hill AJ, et al. Cell-free DNA comprises an in vivo nucleosome footprint that informs its tissues-of-origin. Cell 2016;164:57-68. [Crossref] [PubMed]
- Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001;61:1659-65. [PubMed]
- Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90. [Crossref] [PubMed]
- Sorber L, Zwaenepoel K, Deschoolmeester V, et al. Circulating cell-free nucleic acids and platelets as a liquid biopsy in the provision of personalized therapy for lung cancer patients. Lung Cancer 2017;107:100-7. [Crossref] [PubMed]
- Pérez-Callejo D, Romero A, Provencio M, et al. Liquid biopsy based biomarkers in non-small cell lung cancer for diagnosis and treatment monitoring. Transl Lung Cancer Res 2016;5:455-65. [Crossref] [PubMed]
- Diehl F, Li M, He Y, et al. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods 2006;3:551-9. [Crossref] [PubMed]
- Cai X, Janku F, Zhan Q, et al. Accessing genetic information with liquid biopsies. Trends Genet 2015;31:564-75. [Crossref] [PubMed]
- Worrillow L, Baskaran P, Care MA, et al. An ultra-deep sequencing strategy to detect sub-clonal TP53 mutations in presentation chronic lymphocytic leukaemia cases using multiple polymerases. Oncogene 2016;35:5328-36. [Crossref] [PubMed]
- Baer C, Kern W, Koch S, et al. Ultra-deep sequencing leads to earlier and more sensitive detection of the tyrosine kinase inhibitor resistance mutation T315I in chronic myeloid leukemia. Haematologica 2016;101:830-8. [Crossref] [PubMed]
- Kivioja T, Vähärautio A, Karlsson K, et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods 2011;9:72-4. [Crossref] [PubMed]
- Reguart N, Teixidó C, Giménez-Capitán A, et al. Identification of ALK, ROS1 and RET fusions by a multiplexed mRNA-based assay in formalin-fixed, paraffin-embedded samples from advanced non–small-cell lung cancer patients. Clin Chem 2017;63:751-60. [Crossref] [PubMed]
- Polytarchou C, Oikonomopoulos A, Mahurkar S, et al. Assessment of circulating microRNAs for the diagnosis and disease activity evaluation in patients with ulcerative colitis by using the Nanostring technology. Inflamm Bowel Dis 2015;21:2533-9. [Crossref] [PubMed]
- Wang X, Liu H, Zhao C, et al. The DEAD-box RNA helicase 51 controls non-small cell lung cancer proliferation by regulating cell cycle progression via multiple pathways. Sci Rep 2016;6:26108. [Crossref] [PubMed]
- Alexandrov LB, Jones PH, Wedge DC, et al. Clock-like mutational processes in human somatic cells. Nat Genet 2015;47:1402-7. [Crossref] [PubMed]
- Genovese G, Kahler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 2014;371:2477-87. [Crossref] [PubMed]
- Gormally E, Vineis P, Matullo G, et al. TP53 and KRAS2 mutations in plasma DNA of healthy subjects and subsequent cancer occurrence: a prospective study. Cancer Res 2006;66:6871-6. [Crossref] [PubMed]
- Chae YK, Davis AA, Carneiro BA, et al. Concordance between genomic alterations assessed by next-generation sequencing in tumor tissue or circulating cell-free DNA. Oncotarget 2016;7:65364-73. [PubMed]
- Kuderer NM, Burton KA, Blau S, et al. Comparison of 2 commercially available next-generation sequencing platforms in oncology. JAMA Oncol 2017;3:996-8. [Crossref] [PubMed]
- Mengelbier LH, Karlsson J, Lindgren D, et al. Intratumoral genome diversity parallels progression and predicts outcome in pediatric cancer. Nat Commun 2015;6:6125. [Crossref] [PubMed]
- National Lung Screening Trial Research, Aberle DR, Adams AM, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011;365:395-409. [Crossref] [PubMed]
- Blood Profiling Atlas in Cancer. Part of the U.S. Cancer Moonshot initiative. Available online: https://medium.com/cancer-moonshot/blood-profiling-atlas-in-cancer-21261949bafe#.g45qbm9l9
- Circulating Cell-free Genomic Atlas Study (CCGA). Available online: https://clinicaltrials.gov/ct2/show/NCT02889978
- Arriagada R, Dunant A, Pignon JP, et al. Long-term results of the international adjuvant lung cancer trial evaluating adjuvant cisplatin-based chemotherapy in resected lung cancer. J Clin Oncol 2010;28:35-42. [Crossref] [PubMed]
- Overman MJ, Modak J, Kopetz S, et al. Use of research biopsies in clinical trials: are risks and benefits adequately discussed? J Clin Oncol 2013;31:17-22. [Crossref] [PubMed]
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [Crossref] [PubMed]
- Aupérin A, Le Pechoux C, Rolland E, et al. Meta-analysis of concomitant versus sequential radiochemotherapy in locally advanced non-small-cell lung cancer. J Clin Oncol 2010;28:2181-90. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Novello S, Barlesi F, Califano R, et al. ESMO Guidelines Committee. Metastatic non-small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 2016;27:v1-v27. [Crossref] [PubMed]
- Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 2016;8:346ra92. [Crossref] [PubMed]
- van der Wekken AJ, Saber A, Hiltermann TJ, et al. Resistance mechanisms after tyrosine kinase inhibitors afatinib and crizotinib in non-small cell lung cancer, a review of the literature. Crit Rev Oncol Hematol 2016;100:107-16. [Crossref] [PubMed]
- Paweletz CP, Sacher AG, Raymond CK, et al. Bias-corrected targeted next-generation sequencing for rapid, multiplexed detection of actionable alterations in cell-free DNA from advanced lung cancer patients. Clin Cancer Res 2016;22:915-22. [Crossref] [PubMed]
- Thompson JC, Yee SS, Troxel AB, et al. Detection of therapeutically targetable driver and resistance mutations in lung cancer patients by next-generation sequencing of cell-free circulating tumor DNA. Clin Cancer Res 2016;22:5772-82. [Crossref] [PubMed]


