MET in human cancer: germline and somatic mutations
Deregulated MET signaling occurs in cancer through several mechanisms including overexpression, amplification, autocrine signaling, and mutational activation. In this chapter, we focus on the breadth of MET activating mutations that have been discovered and their tumorigenic consequences. Studies of these MET mutations have been invaluable for our understanding of the tumor initiating activity of MET, receptor tyrosine kinase (RTK) recycling and regulation, and mechanisms of resistance to kinase inhibition. In addition, we will discuss the various tumor types where MET mutations have been found and what these mutations have revealed about the significant role that mutationally-activated MET plays in tumor initiation, progression, and therapeutic resistance.
MET kinase domain mutations in hereditary papillary renal carcinoma (HPRC)
The first activating mutations identified within the MET gene were discovered by a genome-wide analysis of families with HPRC (1). These seminal studies were the first genetic evidence demonstrating oncogenic activity of MET in humans. The germline missense mutations identified in HPRC patients (M1149T, V1206L, V1238I, D1246N, and Y1248C) flank the critical tyrosine residues Y1234 and Y1235 within the kinase domain (Figure 1). In addition, Schmidt et al. discovered somatic missense MET mutations (D1246H, Y1228C, and M1268T) within the same region in sporadic renal carcinomas (1,2). Cytogenetic studies revealed that the papillary renal carcinomas harboring MET mutations also contained trisomy of chromosome 7 (MET is located at 7q31). In each tumor, the Chromosome 7 triplication consisted of the non-random duplication of the chromosome harboring the mutated MET allele (3). The requirement for a second copy of the mutant MET allele in papillary renal carcinomas suggested that there is a necessary dose of activated MET required for tumor initiation in the kidney. Importantly, these findings revealed that mutated MET is a driver gene in hereditary and sporadic papillary renal carcinomas (SPRC).
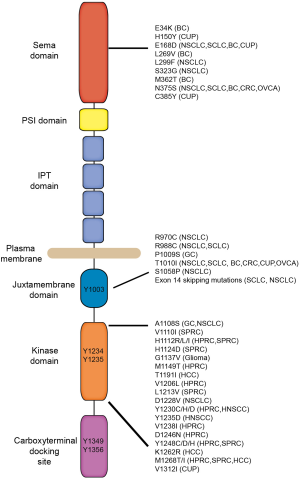
Understanding how mutations within the MET kinase domain affect activation and downstream signaling is vital for our understanding of dysregulated RTK signaling and for the development of effective kinase inhibitors. Several in vitro, xenograft, and transgene experiments verified the inherent oncogenic activity of the MET kinase domain mutations found in HPRC. These studies revealed that the kinase domain Met mutations induce constitutive receptor activation (2,4,5) and mutationally activated Met can be ligand-dependent or ligand-independent (5-8). Knock-in in vivo models of the kinase domain mutations were developed to characterize how mutationally activated MET effects tumorigenesis from initiation to malignant progression. Germline knock-in mouse models were created carrying unique Met kinase mutations including WT, D1226N, Y1228C, M1248T, and M1248T/L1193V (9,10). Interestingly the different mutant Met knock-in lines developed unique tumor profiles including carcinomas, sarcomas, and lymphomas. For example, MetM1248T mice developed a mix of carcinomas and lymphomas while mice harboring D1226N, Y1228C, and M1248T/L1193V mutations developed a high frequency of sarcomas and some lymphomas. These mouse models also replicated the genomic events observed in human HPRC where nonrandom duplication of the mutant Met allele was observed in the majority of the tumors. Even though the knock-in mutation models never developed renal carcinomas, when placed on an FVB/N background, each Met mutant (except for D1226N) developed aggressive mammary carcinomas (11). Again, unique mammary carcinoma phenotypes were observed between the M1248T, Y1228C, and M1248T/L1193V lines (12). Since the only differences between these animals were the Met mutations and the murine background strain, this study indicated that either the mutated kinase structure itself or the level of kinase they impose (or both) influence the tissue-specificity for tumor formation. Overall, these studies demonstrated that the activating mutations affect more than just MET kinase activity and have the potential to drive tumorigenesis in numerous tissue types.
After the discovery of MET mutations in HRPC, studies in other solid tumors identified MET kinase domain mutations and some mutations outside the kinase domain in childhood hepatocellular carcinomas, breast cancer, colorectal cancer (CRC), head and neck squamous cell cancers (HNSCC), gastric carcinomas (GC), and cancers of unknown primary origin (CUP) (Figure 1) (13-21). For several years after the initial discovery, the small number of MET kinase activating mutations identified in other carcinomas suggested that mutations within the MET kinase domain were rare events in cancer. However, recent genomic screens have revealed that activating MET mutations are more frequent than initially thought (22) (COSMIC database at www.cancer.sanger.ac.uk/cosmic). The diversity of cancers in which MET mutations have been identified suggests that mutationally activated MET plays a significant role in the tumorigenic process in a wide range of cell types.
MET juxtamembrane and Sema domain mutations
Since the original screens for kinase domain mutations in other solid cancers identified few variants, searches expanded to regions outside of the kinase domain. In addition to the discovery of novel driver MET mutations, these findings have been critical for our understanding of receptor tyrosine kinase recycling and downregulation. The first juxtamembrane domain (JM) mutations were discovered in a gastric cancer (P1009S) and a breast cancer biopsy (T1010I) (Figure 1) (16). Though the importance and frequency of these mutations was later established in several lung cancer studies (23,24). In a sequencing analysis of small cell lung cancers (SCLC) and non-small cell lung cancers (NSCLC), Ma et al. identified missense mutations in the JM domain (R988C, T1010I, S1058P) and the Sema domain (E168D, L299F, S323G, and N375S) (23,25). A separate study in NSCLC and CRCs identified several mutations in the Sema (N375S) and the JM (R970C and T992I) domains (24). These JM domain mutations were shown to attenuate MET receptor ubiquitination and degradation and prolong MET signaling. Sema domain mutations have not been carefully evaluated but likely affects the structure of the ligand-binding domain (26). In addition, novel intronic mutations flanking exon 14 were discovered that result in an alternatively spliced MET transcript which encodes for a deletion of the JM domain (METex14del) (24,25). Like the JM domain mutations, METex14del receptor downregulation is abrogated by loss of the Cbl site on the JM domain and results in elevated, membrane expression of MET. Importantly, these studies uncovered a novel and distinct mechanism of oncogenic RTK activation through altered RTK downregulation (27,28). The incidence of MET mutations in lung cancers is 3% in squamous cell lung cancers, 5.6% in NSCLC, and 8% in lung adenocarcinomas (29-31). The implications of MET JM domain mutations and receptor regulation on the clinical outcome of lung cancer patients have become evident in recent years (32-37). Moreover, the importance of these Sema and JM domain mutations is not limited to lung cancer. MET mutations have been detected in 9% of advanced breast cancer (20) and 7.4% advanced ovarian cancer patients (38). These studies underscore the role of mutationally activated MET in a wide range of cancers and the diverse mechanisms by which RTKs can achieve oncogenic activity.
Reemergence of MET kinase domain mutations during resistance
Tyrosine kinase inhibitors (TKIs) have had significant success in breast cancer (trastuzumab), melanoma (vemurafenib), and lung cancer (erlotinib), but in spite of these promising results, the clinical response to TKIs is often not durable. The efficacy of targeting kinases has been clearly demonstrated in lung cancer; however these studies have also highlighted the numerous mechanisms by which acquired resistance occurs. The presence of EGFR mutations, ALK fusions, and MET amplification in NSCLC has allowed for the clinical development of several therapeutic approaches using TKIs. For example, EGFR-mutant NSCLCs are sensitive to EGFR TKIs (gefitinib and erlotinib); however, resistance typically develops after 9–14 months (39,40). The most common mechanism of TKI resistance is a second-site mutation (T790M) in the EGFR kinase domain, however in approximately 20% of cases, EGFR inhibition leads to the expansion of subclones harboring amplified MET. Until recently, MET kinase domain mutations have been limited to papillary renal carcinomas, yet the use of TKIs in lung cancer revealed that MET kinase domain mutations may be a mechanism of therapeutic resistance in refractory lung cancer. In a study of a lung adenocarcinoma patient who had progressed on erlotinib treatment, both mutated EGFR and amplified MET were identified in the tumor. Treatment with combined MET and EGFR inhibitors (savolitinib and osimertinib) resulted in a dramatic clinical response (41). When resistance developed to combined MET and EGFR inhibition, a MET D1228V kinase domain mutation was detected. In a separate study, a NSCLC patient with a METex14 deletion was treated with the MET inhibitor crizotinib and upon progression a D1228N mutation was detected (42). Likewise in NSCLC patient with a METex14 deletion, a Y1230C mutation was also detected at a very low frequency (mutant allele frequency =0.3%); however after 13 months of crizotinib treatment the tumor progressed and the Y1230C allele was detected in 3.5% of circulating tumor DNA (43). These results indicate that MET kinase domain mutations may be a mechanism to circumvent MET TKI inhibition. Several earlier studies have demonstrated the ability of MET kinase domain mutations to diminish the efficacy of MET inhibitors (44-46). It is likely that additional and novel mutations will be identified with the emergence of genomic profiling of recurrent disease through biopsies, circulating tumor cells, and circulating DNA. Therefore it is essential that we gain a clear understanding of how MET activating mutations alter the three-dimensional structure of the receptor in order to predict and develop effective MET inhibitors.
Summary
In summary, the search for MET mutations has uncovered the variety of cancers that deregulated MET affects from the early stages of tumor initiation to therapeutic resistance and recurrence. Moreover, the variety of mutations identified within the MET receptor has illuminated unique mechanisms of tumor initiation including nonrandom duplication of mutant alleles, intronic splice site alterations, and altered receptor downregulation. Advances in genomic screening and structural analyses are likely to shed additional light on the prevalence and functional activity of mutationally-activated MET. With this knowledge, MET-targeted therapies may benefit patients in a wide range of cancers.
Acknowledgements
The authors would like to acknowledge Dr. George Vande Woude and Dafna Kaufman for critical reading of the manuscript. We apologize to authors whose studies we were unable to cite because of space restrictions.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Schmidt L, Duh FM, Chen F, et al. Germline and somatic mutations in the tyrosine kinase domain of the Met proto-oncogene in papillary renal carcinomas. Nat Genet 1997;16:68-73. [Crossref] [PubMed]
- Schmidt L, Junker K, Nakaigawa N, et al. Novel mutations of the MET proto-oncogene in papillary renal carcinomas. Oncogene 1999;18:2343-50. [Crossref] [PubMed]
- Zhuang Z, Park WS, Pack S, et al. Trisomy 7-harbouring non-random duplication of the mutant MET allele in hereditary papillary renal carcinomas. Nat Genet 1998;20:66-9. [Crossref] [PubMed]
- Jeffers M, Schmidt L, Nakaigawa N, et al. Activating mutations for the met tyrosine kinase receptor in human cancer. Proc Natl Acad Sci U S A 1997;94:11445-50. [Crossref] [PubMed]
- Jeffers M, Fiscella M, Webb CP, et al. The mutationally activated Met receptor mediates motility and metastasis. Proc Natl Acad Sci U S A 1998;95:14417-22. [Crossref] [PubMed]
- Michieli P, Basilico C, Pennacchietti S, et al. Mutant Met-mediated transformation is ligand-dependent and can be inhibited by HGF antagonists. Oncogene 1999;18:5221-31. [Crossref] [PubMed]
- Wang R, Ferrell LD, Faouzi S, et al. Activation of the Met receptor by cell attachment induces and sustains hepatocellular carcinomas in transgenic mice. J Cell Biol 2001;153:1023-34. [Crossref] [PubMed]
- Jeffers MF, Vande Woude GF. Activating mutations in the Met receptor overcome the requirement for autophosphorylation of tyrosines crucial for wild type signaling. Oncogene 1999;18:5120-25. [Crossref] [PubMed]
- Graveel C, Su Y, Koeman J, et al. Activating Met mutations produce unique tumor profiles in mice with selective duplication of the mutant allele. Proc Natl Acad Sci U S A 2004;101:17198-203. [Crossref] [PubMed]
- Graveel CR, London CA, Vande Woude GF. A mouse model of activating Met mutations. Cell Cycle 2005;4:518-20. [Crossref] [PubMed]
- Graveel CR, DeGroot JD, Su Y, et al. Met induces diverse mammary carcinomas in mice and is associated with human basal breast cancer. Proc Natl Acad Sci U S A 2009;106:12909-14. [Crossref] [PubMed]
- Graveel CR, DeGroot JD, Sigler RE, et al. Germline met mutations in mice reveal mutation- and background-associated differences in tumor profiles. PLoS One 2010;5:e13586. [Crossref] [PubMed]
- Park WS, Dong SM, Kim SY, et al. Somatic mutations in the kinase domain of the Met/hepatocyte growth factor receptor gene in childhood hepatocellular carcinomas. Cancer Res 1999;59:307-10. [PubMed]
- Di Renzo MF, Olivero M, Martone T, et al. Somatic mutations of the MET oncogene are selected during metastatic spread of human HNSC carcinomas. Oncogene 2000;19:1547-55. [Crossref] [PubMed]
- Aebersold DM, Landt O, Berthou S, et al. Prevalence and clinical impact of Met Y1253D-activating point mutation in radiotherapy-treated squamous cell cancer of the oropharynx. Oncogene 2003;22:8519-23. [Crossref] [PubMed]
- Lee JH, Han SU, Cho H, et al. A novel germ line juxtamembrane Met mutation in human gastric cancer. Oncogene 2000;19:4947-53. [Crossref] [PubMed]
- Ghadjar P, Blank-Liss W, Simcock M, et al. MET Y1253D-activating point mutation and development of distant metastasis in advanced head and neck cancers. Clin Exp Metastasis 2009;26:809-15. [Crossref] [PubMed]
- Stella GM, Benvenuti S, Gramaglia D, et al. MET mutations in cancers of unknown primary origin (CUPs). Hum Mutat 2011;32:44-50. [Crossref] [PubMed]
- Liu S, Meric-Bernstam F, Parinyanitikul N, et al. Functional consequence of the MET-T1010I polymorphism in breast cancer. Oncotarget 2015;6:2604-14. [Crossref] [PubMed]
- de Melo Gagliato D, Jardim DL, Falchook G, et al. Analysis of MET genetic aberrations in patients with breast cancer at MD Anderson Phase I unit. Clin Breast Cancer 2014;14:468-74. [Crossref] [PubMed]
- Neklason DW, Done MW, Sargent NR, et al. Activating mutation in MET oncogene in familial colorectal cancer. BMC Cancer 2011;11:424. [Crossref] [PubMed]
- Vigna E, Comoglio PM. Targeting the oncogenic Met receptor by antibodies and gene therapy. Oncogene 2015;34:1883-9. [Crossref] [PubMed]
- Ma PC, Kijima T, Maulik G, et al. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res 2003;63:6272-81. [PubMed]
- Kong-Beltran M, Seshagiri S, Zha J, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res 2006;66:283-9. [Crossref] [PubMed]
- Ma PC, Jagadeeswaran R, Jagadeesh S, et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res 2005;65:1479-88. [Crossref] [PubMed]
- Kong-Beltran M, Stamos J, Wickramasinghe D. The Sema domain of Met is necessary for receptor dimerization and activation. Cancer Cell 2004;6:75-84. [Crossref] [PubMed]
- Peschard P, Fournier TM, Lamorte L, et al. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol Cell 2001;8:995-1004. [Crossref] [PubMed]
- Mak HH, Peschard P, Lin T, et al. Oncogenic activation of the Met receptor tyrosine kinase fusion protein, Tpr-Met, involves exclusion from the endocytic degradative pathway. Oncogene 2007;26:7213-21. [Crossref] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014;511:543-50. [Crossref] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012;489:519-25. [Crossref] [PubMed]
- Heist RS, Shim HS, Gingipally S, et al. MET Exon 14 Skipping in Non-Small Cell Lung Cancer. Oncologist 2016;21:481-6. [Crossref] [PubMed]
- Schrock AB, Frampton GM, Suh J, et al. Characterization of 298 Patients with Lung Cancer Harboring MET Exon 14 Skipping Alterations. J Thorac Oncol 2016;11:1493-502. [Crossref] [PubMed]
- Tong JH, Yeung SF, Chan AW, et al. MET Amplification and Exon 14 Splice Site Mutation Define Unique Molecular Subgroups of Non-Small Cell Lung Carcinoma with Poor Prognosis. Clin Cancer Res 2016;22:3048-56. [Crossref] [PubMed]
- Awad MM, Oxnard GR, Jackman DM, et al. MET Exon 14 Mutations in Non-Small-Cell Lung Cancer Are Associated With Advanced Age and Stage-Dependent MET Genomic Amplification and c-Met Overexpression. J Clin Oncol 2016;34:721-30. [Crossref] [PubMed]
- Jorge SE, Schulman S, Freed JA, et al. Responses to the multitargeted MET/ALK/ROS1 inhibitor crizotinib and co-occurring mutations in lung adenocarcinomas with MET amplification or MET exon 14 skipping mutation. Lung Cancer 2015;90:369-74. [Crossref] [PubMed]
- Paik PK, Drilon A, Fan PD, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov 2015;5:842-9. [Crossref] [PubMed]
- Zheng D, Wang R, Ye T, et al. MET exon 14 skipping defines a unique molecular class of non-small cell lung cancer. Oncotarget 2016;7:41691-702. [PubMed]
- Tang C, Jardim DL, Falchook GS, et al. MET nucleotide variations and amplification in advanced ovarian cancer: characteristics and outcomes with c-Met inhibitors. Oncoscience 2013;1:5-13. [PubMed]
- Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med 2009;361:947-57. [Crossref] [PubMed]
- Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009;361:958-67. [Crossref] [PubMed]
- Bahcall M, Sim T, Paweletz CP, et al. Acquired METD1228V Mutation and Resistance to MET Inhibition in Lung Cancer. Cancer Discov 2016;6:1334-41. [Crossref] [PubMed]
- Heist RS, Sequist LV, Borger D, et al. Acquired Resistance to Crizotinib in NSCLC with MET Exon 14 Skipping. J Thorac Oncol 2016;11:1242-5. [Crossref] [PubMed]
- Ou SI, Young L, Schrock AB, et al. Emergence of Preexisting MET Y1230C Mutation as a Resistance Mechanism to Crizotinib in NSCLC with MET Exon 14 Skipping. J Thorac Oncol 2017;12:137-40. [Crossref] [PubMed]
- Berthou S, Aebersold DM, Schmidt LS, et al. The Met kinase inhibitor SU11274 exhibits a selective inhibition pattern toward different receptor mutated variants. Oncogene 2004;23:5387-93. [Crossref] [PubMed]
- Zimmer Y, Vaseva AV, Medova M, et al. Differential inhibition sensitivities of MET mutants to the small molecule inhibitor SU11274. Cancer Lett 2010;289:228-36. [Crossref] [PubMed]
- Medova M, Pochon B, Streit B, et al. The novel ATP-competitive inhibitor of the MET hepatocyte growth factor receptor EMD1214063 displays inhibitory activity against selected MET-mutated variants. Molecular cancer therapeutics 2013;12:2415-24. [Crossref] [PubMed]


