One size doesn’t fit all: unraveling the diversity of factors and interactions that drive E. coli urovirulence
Uropathogenic Escherichia coli (UPEC): a common pathogen without a clear definition
Urinary tract infections (UTIs) are common (with 11 million cases annually in the USA), costly (with $5 billion spent per year), and increasingly antibiotic resistant (1). The clinical manifestations and symptomatologies of UTIs are strikingly complex and result from the interactions between diverse uropathogens and host urothelial tissues. The most common cause of UTIs is UPEC, which cause up to 80% of community-acquired UTIs and 65% of hospital-acquired UTIs (1). Despite appropriate treatment for the initial infection, UTIs are frequently recurrent as 25–35% of initial infections are followed by a recurrent UTI (rUTI) (2). In addition, the high level of same-strain recurrences, in which a single bacterial strain (as defined by genetic identity) causes two or more consecutive UTIs, strongly suggests that there exist non-pathogenic reservoirs within hosts (2). One established reservoir is the gastrointestinal tract (GIT) microbiota (3). UPEC strains can maintain non-pathogenic colonization of the GIT, where they form a reservoir and subsequently seed extraintestinal infections such as UTI. In this review, we detail critical gaps in our understanding of the pathogenic and non-pathogenic colonization of UPEC in different habitats in the host and describe a new perspective on UTI susceptibility that better reflects the complexity of this disease.
In contrast to other Escherichia coli (E. coli) pathotypes, UPEC are distinct in their lack of a defined set of genes that distinguish them from non-UPEC (4). In other E. coli pathotypes, virulence is demarcated by a clearly defined set of virulence genes, such as the stx gene in Shiga-toxin producing E. coli (5). This likely reflects the broad definition of UPEC as any strain that is recovered from the urine of symptomatic UTI patients, a classification that fails to account for differences in host susceptibility or the possibility for multiple evolutionary and mechanistic paths to urovirulence. Further, despite widespread acceptance that the gut is a reservoir of UPEC that lead to UTI (3,6), we know very little about the host and bacterial factors that promote the establishment and maintenance of UPEC in this habitat. Towards the goal of identifying conserved bacterial features enabling urovirulence and gut colonization, recent work has integrated different animal models with comparative genomics and transcriptomics to test the long-held view that the ability of E. coli to cause UTIs correlates with carriage of putative urovirulence factors (PUFs) and that pathogenicity is restricted to certain branches of the E. coli phylogenetic tree (7). This work, coupled with the diversity of urine isolates observed clinically, has led us to a new way of thinking about UTI—a “Key and Lock” model. This model posits that the outcome of an encounter between a specific host and potential UPEC isolate is dependent upon the combination of the particular fitness state of the UPEC isolate (the “key”) matched with the particular host environment and susceptibility (the “lock”). Below we describe what is being learned about each of these components.
The host “Lock”: UTI susceptibility is shaped by host genetic, environmental and behavioral factors
Epidemiological studies have shown that host susceptibility is variable and mediated by numerous genetic, environmental, and behavioral risk factors (8). Many of these risk factors have been investigated in experimental mouse infection models. Mice have similar bladder morphology and express many of the same surface proteins as humans. Mice also respond to infection with phenotypes that recapitulate clinical findings in humans [e.g., epithelial exfoliation, cytokines, pyuria, and antimicrobial peptide, as reviewed in (9) and (10)]. However, as with all animal models of any human infectious disease, a single animal model using a host with a single genetic background cannot possibly reflect all of the variation observed in clinical UTIs driven by diverse genetic, behavioral and environmental differences within human populations. Thus, multiple animal models of UTI have been developed, each reflecting a portion of UTI clinical complexity (Table 1). These different mouse models have been critical in defining particular host-pathogen interactions that contribute to UTI in both mice and humans as well as elucidating unappreciated aspects of clinical UPEC infection. However, while investigations of UPEC pathogenesis in mice have yielded important insights into the clinical progression of UTI in humans, such studies have relied almost exclusively on model UPEC strains, such as UTI89 and CFT073, which represent only a small section of the overall E. coli phylogenetic tree despite the remarkable diversity of UPEC strains in the clinic (7). Thus, it is unknown whether diverse UPEC strains found in the clinic follow the same pathogenic pathways or how these diverse UPEC interact with different host susceptibility factors.

Full table
In addition to host factors that directly impact susceptibility to bladder colonization by UPEC, recent advances in the understanding of the gut microbiota suggest that host factors influence the composition of the UPEC reservoir in the gut, thus affecting host susceptibility to both acute and rUTIs. In healthy adults, E. coli account for ~0.8% of the total microbiota and are the primary facultative anaerobe in the GIT microbial community (23). While the majority of adults carry E. coli asymptomatically in their gut, blooms of E. coli are associated with a number of intestinal diseases including diarrheal diseases, inflammatory bowel diseases (IBD) such as Crohn’s disease, and colorectal cancer (24,25). One prominent host factor that likely influences the E. coli population in the gut is inflammation, which can be triggered in numerous ways including infection by a pathogen, an imbalance of the existing microbiota, or exposure to antibiotics during clinical intervention (26,27). Interestingly, biopsy specimens from patients with Crohn’s disease and ulcerative colitis, two IBD syndromes, revealed that these patients have a 3–4 log increase in the levels of Enterobacteriaceae in their intestines over healthy controls, with a significant increase in the abundance of E. coli from the B2 and D phylogenetic groups (24). This has also been observed in conventionally raised mice treated with the antibiotic streptomycin or dextran sulfate sodium (DSS), a chemical used to superficially stimulate Crohn’s disease in murine animals (28). Of interest, several clinical studies have found that patients with intestinal inflammatory diseases, like IBD and AIDS, have an increased incidence of rUTI (29,30). Together, these data suggest that intestinal inflammation may alter conditions in the gut in a way that enhances UPEC colonization, ultimately increasing the likelihood of a downstream UTI, particularly in individuals that are susceptible to chronic/recurrent cystitis.
The UPEC “Key”: dynamic bacterial features link UPEC colonization of the gut and bladder
As a species, E. coli strains possess striking genetic diversity and two strains can differ in their gene content by up to 40% (31). As such, individual genes can be classified as “core genes” belonging to all sequenced E. coli genomes or “variable” genes, those that are only found in some, but not all strains (32). Analysis of the genetic diversity in these core genes can be used to subdivide E. coli strains into a phylogenetic tree with strains grouping into different clades, including clades A, B1, B2, D and E that are commonly associated with humans (33). Using a PCR-based, fragment analysis of largely conserved genomic loci, molecular epidemiology studies have shown that, in the western world, between 50% and 75% of UPEC isolates originate from clade B2 (7,34).
Many studies have identified PUF genes and described them as being enriched in UPEC isolates relative to E. coli isolates found in the gut. PUF genes are often posited to increase the urovirulence of UPEC strains (35). However, no study has ever identified a single, core set of genes that clearly discriminate a UPEC strain from a non-UPEC strain and the role of many PUF genes in bladder persistence and colonization remains unknown (4). B2 strains of E. coli are commonly used in the study of UTIs and typically encode many PUFs in their genomes; however, approximately 25–50% of UPEC UTIs are caused by strains outside of clade B2 (non-B2 strains) that carry few PUF genes. To address this disconnect between model B2 strains and the diversity of UPEC seen clinically, recent work used systematic genetic, transcriptional, and phenotypic comparisons among dozens of diverse UPEC clinical isolates to examine the complexity of uropathogenicity (7). This interdisciplinary study showed that PUFs were associated with strains of the B2 clade whether isolated from the urine of UTI patients or other sources. Further, it revealed that transcriptional and phenotypic features of E. coli isolates were better predictors of virulence in defined model systems of acute cystitis than the carriage of PUFs. This work suggested that differential expression and/or regulation of conserved functions is a key driver of UPEC pathogenesis. Further, while PUFs may be important factors in different, sometimes unknown, aspects of UPEC virulence, this study proposes that there may be more than one “pathway” or one set of genes/mechanisms that allow E. coli to cause disease in the bladder similar to what has been shown for different E. coli pathotypes causing disease in the GIT [as reviewed in (5)]. Thus, we argue that to more fully understand human UTI, studies elucidating the factors and mechanisms underlying UPEC urovirulence must be broadened to include a more diverse set of strains and extended to compare not only patterns of gene carriage but also of gene expression and key phenotypes among these strains.
In addition to our relatively shallow understanding of bladder colonization and persistence by genetically diverse UPEC, the bacterial factors that influence colonization and persistence of UPEC in the gut are even less clear. However, several studies have shown that at the time of UTI, the dominant E. coli strain in the urine and feces of an infected woman is the same (3,6). In women with rUTI, this dominant strain can change from one infection to the next or stay the same (3). When strain replacement does occur in women with rUTI, a recent study found that the new E. coli strain is associated with increased fitness in both the gut and bladder, as measured by its ability to colonize these habitats compared to its predecessor (3). Prior to this finding, the long-standing hypothesis regarding UPEC fitness in the gut was that UPEC followed the source-sink model, which posits a fitness tradeoff for UPEC between the “source”, the gut, and the “sink”, the urinary tract, compartments (36). The recent data does not preclude the presence of UPEC genetic factors that provide enhanced fitness in one habitat while lowering fitness in the other, but strongly opposes the idea that UPEC must undergo a complete loss of fitness in one habitat, to be fit in the other. Overall, findings indicate that some UPEC-microbiota interactions in the gut that impact the ability of UPEC to colonize this habitat may also increase fitness in the bladder. Thus, seeding effects between the gut and urinary tract can lead to one dominant UPEC strain emerging across both habitats during a UTI episode. This has implications for our understanding of genetically diverse UPEC, as PUFs, which do not enhance bladder persistence and colonization directly may actually enhance overall urovirulence by enhancing UPEC gut colonization. Ultimately, these studies underscore the necessity of investigating UPEC within the broader context of their human lifestyles—as both gut commensals and genetically diverse opportunistic pathogens.
Matching the “Key” to the “Lock”: the outcome of encounters between a particular host and a particular pathogen reflect the “fit” between the two
Research to date has lead us to a new perspective on UPEC pathogenesis in which UTI risk is determined by diverse bacterial virulence phenotypes, variable and changing host susceptibilities, and the interactions of these phenotypes and susceptibilities in specific host-pathogen combinations. Thus, we propose a multi-dimensional and dynamic “Lock and Key” model of UTI risk in which the “Lock” of host susceptibility is matched against the “Key” of an individual E. coli isolate’s virulence potential (7). Given the multi-dimensionality of this concept, it is best visualized as landscape of interactions between hosts and UPEC (Figure 1). In this model, some UPEC “keys” may act as “master keys” being able to open a variety of different “locks” and some “keys” may be host specific, only functioning on a narrow spectrum of host “locks”. Similarly, some “locks” may differentially accommodate the fit of a broad spectrum of different “keys”, while some may be specific to only a few. Importantly, we posit that not all UPEC “keys” are limited in their activity of colonization and persistence within the urinary tract during an acute infection. Instead, additional attention must be paid to factors that contribute to the ability of UPEC strains to persist within host reservoirs outside of the bladder, such as in the gut microbiota. By incorporating previously unaccounted for phenotypic and transcriptional variation among E. coli strains and their differential responses to dynamic host environments, this conceptual model helps to explain UPEC diversity in the clinic, including the lack of a clear genetic signature of uropathogenesis. However, picturing the “locks and keys” in this analogy as immutable objects that are in incapable of change oversimplifies the state of UTI risk. Instead we suggest that this is better envisioned with both the host susceptibility “lock” and the bacterial pathogen “key” being malleable in nature and changing over time in response to differing conditions due to the dynamic nature of both host and bacterial phenotypes. This is exemplified by recent work showing that UPEC alter their transcriptional expression of attachment organelles from type 1 pili to FML pili during the transition from acute cystitis to chronic cystitis in mouse bladders (37). Notably, this change in expression coincides with inflammation in the mouse bladder epithelium that results in the expression of new ligands that are recognized by newly expressed FML pili, thus enabling the persistence of UPEC that would otherwise be removed during bladder remodeling.
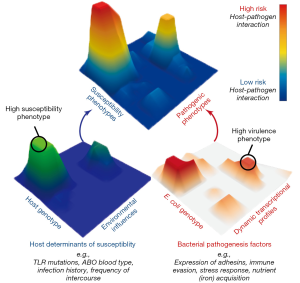
Numerous clinical infections are caused by genetically diverse pathogens that infect hosts with varying levels of susceptibility. Thus, the dynamic “Lock and Key” model of UTI risk is likely translatable to other infectious diseases that lack clear bacterial genetic signatures for pathogenicity. Thus, integrated research methods that combine clinical research with experimental model systems are needed to probe the effects of: (I) host and pathogen genetic diversity; (II) host and bacterial responses to changing infection conditions; (III) infection dynamics that lead to differential susceptibilities and outcomes of bacterial infections in humans and; (IV) pathogen transmission between varied reservoir and infection habitats. This research paradigm promises to yield new insight into the conserved and targetable mechanisms of virulence, critical for development of novel therapeutic strategies that are increasingly needed to face the rising tide of antibiotic resistance.
Acknowledgements
We thank members of the S.J.H. laboratory for their helpful suggestions and insightful comments.
Funding: HL Schreiber IV was supported in part by a 2013 Monsanto Excellence Fund Graduate Fellowship and a 2012 Lucille P. Markey Pathway award. This project was funded by NIH, specifically the NIDDK grant number RO1 DK051406 and the ORWH SCOR grant number P50 DK064540 to SJ Hultgren, and the NIAID grant number U19AI110818 to the Broad Institute.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Disclaimer: The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
References
- Foxman B. Urinary tract infection syndromes: occurrence, recurrence, bacteriology, risk factors, and disease burden. Infect Dis Clin North Am 2014;28:1-13. [Crossref] [PubMed]
- Foxman B, Gillespie B, Koopman J, et al. Risk factors for second urinary tract infection among college women. Am J Epidemiol 2000;151:1194-205. [Crossref] [PubMed]
- Chen SL, Wu M, Henderson JP, et al. Genomic diversity and fitness of E. coli strains recovered from the intestinal and urinary tracts of women with recurrent urinary tract infection. Sci Transl Med 2013;5:184ra60. [Crossref] [PubMed]
- Köhler CD, Dobrindt U. What defines extraintestinal pathogenic Escherichia coli? Int J Med Microbiol 2011;301:642-7. [Crossref] [PubMed]
- Croxen MA, Law RJ, Scholz R, et al. Recent advances in understanding enteric pathogenic Escherichia coli. Clin Microbiol Rev 2013;26:822-80. [Crossref] [PubMed]
- Moreno E, Andreu A, Pigrau C, et al. Relationship between Escherichia coli strains causing acute cystitis in women and the fecal E. coli population of the host. J Clin Microbiol 2008;46:2529-34. [Crossref] [PubMed]
- Schreiber HL 4th, Conover MS, Chou WC, et al. Interplay between dynamic bacterial virulence phenotypes of E. coli and host susceptibility determines UTI risk. Sci Transl Med. In Press 2017.
- Scholes D, Hooton TM, Roberts PL, et al. Risk factors for recurrent urinary tract infection in young women. J Infect Dis 2000;182:1177-82. [Crossref] [PubMed]
- Carey AJ, Tan CK, Ipe DS, et al. Urinary tract infection of mice to model human disease: Practicalities, implications and limitations. Crit Rev Microbiol 2016;42:780-99. [PubMed]
- Barber AE, Norton JP, Wiles TJ, et al. Strengths and Limitations of Model Systems for the Study of Urinary Tract Infections and Related Pathologies. Microbiol Mol Biol Rev 2016;80:351-67. [Crossref] [PubMed]
- Anderson GG, Palermo JJ, Schilling JD, et al. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 2003;301:105-7. [Crossref] [PubMed]
- Rosen DA, Hooton TM, Stamm WE, et al. Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med 2007;4:e329. [Crossref] [PubMed]
- Hannan TJ, Mysorekar IU, Hung CS, et al. Early severe inflammatory responses to uropathogenic E. coli predispose to chronic and recurrent urinary tract infection. PLoS Pathog 2010;6:e1001042. [Crossref] [PubMed]
- O'Brien VP, Hannan TJ, Yu L, et al. A mucosal imprint left by prior Escherichia coli bladder infection sensitizes to recurrent disease. Nat Microbiol 2016;2:16196. [Crossref] [PubMed]
- Schlager TA, LeGallo R, Innes D, et al. B cell infiltration and lymphonodular hyperplasia in bladder submucosa of patients with persistent bacteriuria and recurrent urinary tract infections. J Urol 2011;186:2359-64. [Crossref] [PubMed]
- Song J, Bishop BL, Li G, et al. TLR4-initiated and cAMP-mediated abrogation of bacterial invasion of the bladder. Cell Host Microbe 2007;1:287-98. [Crossref] [PubMed]
- Ragnarsdóttir B, Lutay N, Grönberg-Hernandez J, et al. Genetics of innate immunity and UTI susceptibility. Nat Rev Urol 2011;8:449-68. [Crossref] [PubMed]
- Hawn TR, Scholes D, Li SS, et al. Toll-like receptor polymorphisms and susceptibility to urinary tract infections in adult women. PLoS One 2009;4:e5990. [Crossref] [PubMed]
- Schwartz DJ, Conover MS, Hannan TJ, et al. Uropathogenic Escherichia coli superinfection enhances the severity of mouse bladder infection. PLoS Pathog 2015;11:e1004599. [Crossref] [PubMed]
- Hannan TJ, Roberts PL, Riehl TE, et al. Inhibition of Cyclooxygenase-2 Prevents Chronic and Recurrent Cystitis. EBioMedicine 2014;1:46-57. [Crossref] [PubMed]
- Flores-Mireles AL, Pinkner JS, Caparon MG, et al. EbpA vaccine antibodies block binding of Enterococcus faecalis to fibrinogen to prevent catheter-associated bladder infection in mice. Sci Transl Med 2014;6:254ra127. [Crossref] [PubMed]
- Flores-Mireles AL, Walker JN, Bauman TM, et al. Fibrinogen Release and Deposition on Urinary Catheters Placed during Urological Procedures. J Urol 2016;196:416-21. [Crossref] [PubMed]
- Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012;486:207-14. [Crossref] [PubMed]
- Kotlowski R, Bernstein CN, Sepehri S, et al. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut 2007;56:669-75. [Crossref] [PubMed]
- Arthur JC, Perez-Chanona E, Mühlbauer M, et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012;338:120-3. [Crossref] [PubMed]
- Stecher B, Robbiani R, Walker AW, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol 2007;5:2177-89. [Crossref] [PubMed]
- Winter SE, Lopez CA, Bäumler AJ. The dynamics of gut-associated microbial communities during inflammation. EMBO Rep 2013;14:319-27. [Crossref] [PubMed]
- Lupp C, Robertson ML, Wickham ME, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe 2007;2:119-29. [Crossref] [PubMed]
- Kyle J. Urinary complications of Crohn's disease. World J Surg 1980;4:153-60. [Crossref] [PubMed]
- Evans JK, McOwan A, Hillman RJ, et al. Incidence of symptomatic urinary tract infections in HIV seropositive patients and the use of cotrimoxazole as prophylaxis against Pneumocystis carinii pneumonia. Genitourin Med 1995;71:120-2. [PubMed]
- Welch RA, Burland V, Plunkett G 3rd, et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc Natl Acad Sci U S A 2002;99:17020-4. [Crossref] [PubMed]
- Vernikos G, Medini D, Riley DR, et al. Ten years of pan-genome analyses. Curr Opin Microbiol 2015;23:148-54. [Crossref] [PubMed]
- Rasko DA, Rosovitz MJ, Myers GS, et al. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol 2008;190:6881-93. [Crossref] [PubMed]
- Ejrnæs K, Stegger M, Reisner A, et al. Characteristics of Escherichia coli causing persistence or relapse of urinary tract infections: phylogenetic groups, virulence factors and biofilm formation. Virulence 2011;2:528-37. [Crossref] [PubMed]
- Luo Y, Ma Y, Zhao Q, et al. Similarity and divergence of phylogenies, antimicrobial susceptibilities, and virulence factor profiles of Escherichia coli isolates causing recurrent urinary tract infections that persist or result from reinfection. J Clin Microbiol 2012;50:4002-7. [Crossref] [PubMed]
- Sokurenko EV, Gomulkiewicz R, Dykhuizen DE. Source-sink dynamics of virulence evolution. Nat Rev Microbiol 2006;4:548-55. [Crossref] [PubMed]
- Conover MS, Ruer S, Taganna J, et al. Inflammation-Induced Adhesin-Receptor Interaction Provides a Fitness Advantage to Uropathogenic E. coli during Chronic Infection. Cell Host Microbe 2016;20:482-92. [Crossref] [PubMed]


