Neoantigen-based cancer immunotherapy
Introduction
Cancer immunotherapy is now offering opportunities to produce substantial clinical benefits. Recent successes and momentum in immunotherapeutic strategies have shifted the paradigm of patient care and put immunotherapy at the forefront to become the fourth pillar of the standard of care following surgery, chemotherapy and radiotherapy.
In particular, the clinical relevance of T cells in the control of a diverse set of human cancers is now clearly established. However, the nature of antigens (Ags) that allow the immune system to distinguish benign cells from tumor cells has long been unclear and tumor regression Ags remain obscure.
Research efforts in last decades have provided clear evidence that human tumor cells express antigenic determinants (epitopes) that can be recognized by the patients’ autologous T cells. The short peptides that lead to such specific recognition and elimination of cancer cells are presented on the human leukocyte antigen (HLA) molecules and are named the immunopeptidome. CD4+ and CD8+ T lymphocytes have been shown to target epitopes arising from epigenetic, transcriptional, translational and post-translational alterations of tumor cells (1), and up to date, shared tumor-associated Ags (TAAs) have been extensively exploited for therapeutic purposes [e.g., vaccines and adoptive cell therapy (ACT) with gene-engineered T cells], with encouraging, yet controversial, results.
More recently, technological breakthroughs have shown that numerous endogenous mutated cancer proteins, a hallmark of tumor cells, can be processed into peptides and presented on the surface of tumor cells, leading to their immune recognition in vivo as “non-self” or foreign. Targeting such highly specific neoantigens (neoAgs) would enable immune cells to distinguish cancerous from normal cells, avoiding the risk of autoimmunity.
Recent exciting results demonstrated T-cell reactivities against neoAgs in mice (2-4) and in humans (5-8), in both the CD8+ and CD4+ T cell compartments (9-11). Importantly, as neoAgs are exclusively tumor-specific, central T-cell tolerance is not a concern. NeoAg-specific T cells were indeed shown to possess functional avidity that is reaching the avidity strength of anti-viral T cells (12). In contrast, T-cell reactivity toward self-Ags is lower by definition and is achieved only when tolerance to these Ags is not fully developed. Indeed, in cancer patients, TAAs were shown to be recognized in patients by a repertoire of T cells with relatively reduced functional avidity (6). In addition, T-cell responses against neoAgs are not expected to result in autoimmune toxicity against healthy tissues, making therapeutic vaccination with neoAgs highly attractive. However, neoAgs are in large part patient-specific, since the individual mutations found in any pair of tumors are largely distinct (13). Thus, based on current knowledge, it is unlikely that a vaccine can be designed to target shared neoAgs in a large group of patients.
As discussed above, neoAg-reactive T cells were identified in several human cancers including melanoma (5,6), leukemia (14), ovarian cancer (OC) (7) and cholangiocarcinoma (11). Furthermore, interestingly, in patients with melanoma or non-small cell lung carcinoma, the mutational load correlated to the clinical outcome following immunotherapy with anti-CTLA-4 and anti-PD-1, respectively (8,15) and the frequency of neoAg-specific T cells increased in responding patients after therapy. Also, neoAg-reactive CD4+ and CD8+ T cells, already identified in few cancer types, correlated with favorable clinical outcome (5-7,10,11,14). Altogether, these emerging data indicate that neoAg recognition is a major factor in the activity of clinical immunotherapies.
Massively parallel sequencing can now reveal the mutational spectrum of individual tumors (mutanome) with an unprecedented precision and speed (16). These deep sequencing analyses have revealed that solid tumors harbor usually between fifty and thousands of somatic mutations, most of which differ among tumor specimens even within the same tumor type (17). Such sequencing data holds promise as a long-awaited ‘gold mine’ for the identification of unique targets to be exploited in order to design distinct personalized immunotherapy programs aiming to induce, boost or reinvigorate mutation-specific adaptive immunity (18-20).
Current strategies for neo-epitope identification
In the post-genome era, technologies as well as computational tools have emerged that allow the identification of the mutational spectrum of individual tumors (i.e., the mutanome). Upon identification of nonsynonymous mutations, neo-epitopes can be identified, on a patient-specific basis, by several means. Most commonly, neural network algorithms, such as NetMHC, are used to in silico predict high-affinity neo-epitopes derived from mutated sequences (genes) that bind patients own HLA class I molecules (2,3). In silico predicted peptides are then synthesized and used to interrogate patient’s immunity, as described below. In this context, long peptides or mRNA encoding for the mutations can be used (5,10). This approach, called reverse identification, is generating a list of candidate neo-epitopes that can potentially be further filtered on the basis of distinct bioinformatic tools e.g., stability, processing by the immunoproteasome, etc. To date, only HLA Class I, but not Class II restricted peptides can be predicted since the accuracy of prediction of neural network algorithms for CD4 T-cell epitopes remains limited (21,22). Another drawback of the reverse identification approach is that it remains unknown whether neo-epitopes are indeed presented by tumor cells.
In an alternative approach, the HLA ligandome of tumor cells is analyzed. This strategy, called direct identification, requires the elution of the peptides from HLA molecules derived from the tumor tissue of the patient, followed by reversed phase HPLC fractionation and mass spectrometry (MS). Interestingly, although direct identification still needs to be validated by exome and transcriptome sequencing data, MS-based identification of neo-epitopes allows the identification of both CD8+ and CD4+ T-cell neo-epitopes, unlike reverse identification. Therefore, although the sensitivity of the neoAg identification by direct identification remains to be improved, it will most likely represent a key tool in the armamentarium of antigen discovery in the future.
Experimental cellular validation of neoAg antigenicity
A large diversity of bioinformatics and biochemical tools are available to in silico predict, filter or experimentally validate candidate peptides on the basis of their processing, as well as HLA binding affinity and stability. Still, the ultimate demonstration of their potential relevance relies on the experimental validation of their immunogenicity using patient’s own T cells. Recently, several methodologies were developed to interrogate patient’s cellular immunity. These T cell-based functional assays (Figure 1) use either in silico predicted short peptides, exclusively for CD8+ T cells, or long peptides and mRNA to agnostically interrogate both CD4+ and CD8+ T cells (2,3,5,10,23). The selection of short HLA class I restricted peptides is mainly based on predictions from artificial neural networks, as discussed above, and successfully lead to the identification of antigenic neo-epitopes in several studies. Different strategies were established to identify neo-epitopes derived from short, HLA class I-restricted, peptides. These included, on one hand, the direct identification using peptide-HLA multimer complexes (6) and, on the other hand, functional assays (5). In the case where CD4+ or CD8+ T-cell epitopes were identified by MS (direct identification), all of the above strategies (Figure 1) would potentially apply, while their relative sensitivity remains to be determined.
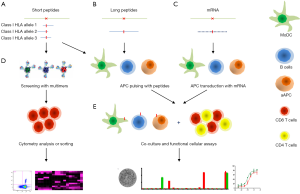
Interestingly, several studies have shown that, in addition to CD8+ T cells, neoAg-specific CD4+ T cells are frequently detected and that their induction after immunization can lead to tumor control, as well as antigen spreading in murine models (9). Several experimental strategies are now used to interrogate the CD4+ T cell responses against the mutanome. These include pulsing with peptides of various lengths, as well as with mRNA. Indeed, a reliable strategy to identify immunogenic T cell neoAgs consists in transfecting autologous antigen presenting cells (APCs), e.g., dendritic cells (DC) or immortalized B cells, with RNA or DNA encoding a sequence of a mutated gene encompassing about 12 residues up and down stream of the point mutation (Figure 1). The transfected APCs are then incubated with patient’s T cells, i.e., tumor infiltrating lymphocytes (TILs) or peripheral blood mononuclear cells (PBMC). Responding T cells can also be propagated upon re-stimulation with Ags-coding APCs. An inherent challenge to this strategy is the definition of the minimal length of T cell epitopes and their MHC restrictions. One way to identify these is by pulsing APCs with synthetic peptides predicted to bind to given HLA molecules by means of in silico algorithms. To unambiguously define the MHC restriction, APCs expressing only one MHC molecule need to be used. In view of the diversity of MHC molecules and processing possibilities of such sequences, it can be tedious to identify the minimal length of T cell epitopes. Moreover, the use of patient’s T cells for neo-epitope T cell discovery is reliable, but requires significant numbers of T cells, which often are limited, and also require massive T-cell expansion that is prone to shifts in their clonal composition (24).
Promises and challenges
Landmark preclinical studies addressed key questions arising from the concept of personalized immunotherapy, by defining pipelines for the identification of immunogenic tumor mutations, by proving their reliability and consistency and by showing a tumor survival benefit upon neoAg vaccination, using peptides and RNA, in both prophylactic and therapeutic settings (2-4,9,25). On this basis, cancer vaccines actively targeting neoAgs have already entered the clinic. Several phase I/II clinical trials are currently on-going, encompassing different strategies including poly-epitopic RNA and peptide vaccines based on high-throughput sequencing (HTS) and in silico prediction, manufactured on demand in metastatic melanoma (NTC02035956, NCT01970358), peptide vaccines based on HTS data combined with MS data in glioblastoma (NCT02149225) and polyepitope plasmid DNA and RNA vaccines in triple-negative breast cancer (NCT 02348320, NCT 02316457).
On the other hand, pivotal work by many groups proved that mutanome-directed T cells represent a major component of TILs that are expanded ex vivo and used for ACT in melanoma and epithelial cancer patients (5,11,12,23). In addition to showing that neoAgs can drive tumor rejection, these studies also triggered a further development in the field of personalized strategies, based on passive as opposed to active immunotherapy. Once identified, neoAg-specific T cells can be isolated from tumors and peripheral blood (26) and specifically expanded to then be re-infused into the patients. An alternative approach envisions using gene-modified T lymphocytes redirected by neoAg-specific T-cell receptors (TCRs) and chimeric Ag receptors (CARs). The safety of the administration of peripheral blood lymphocytes, transduced ex vivo with two different CARs targeting a mutated variant of EGFR (EGFRIIIv), expressed in about 30% of glioblastoma multiform patients, is being currently assessed (NCT01454596). Of note, ACT of a >95% pure population of naturally occurring CD4+ TILs, specific for the ERBB21P neoAg, isolated in a cholangiocarcinoma patient, induced tumor regression (11).
Despite being reliable and extremely promising, personalized immunotherapy targeting unique mutations is facing many technical and logistical hurdles. A major challenge derives from the fact that almost all such mutations are “private”, i.e., unique to a specific tumor. This means that immunogenic mutations must be identified for each patient individually and then experimentally validated, which requires the development of high-throughput and robust methodologies with high fidelity, to both identify mutations and interrogate patient’s T cells in a timely manner for therapeutic applications. Great efforts are currently being made to process the huge amount of data on mutant Ags and identify biomarkers (e.g., gene expression levels, number of predicted peptides per mutation, peptide biochemical properties) disclosing the immunogenic mutations and prioritizing clinically-relevant immunogenic neoAgs. In the future, in silico analyses relying exclusively on DNA/RNA sequencing data will potentially overcome the need for time- and cell-consuming predicted peptide screening. Another key question is how to select the mutations that should be targeted. Driver mutations, as opposed to passenger ones, are involved in the tumorigenesis process and they are critical for tumor cell growth and survival. Thus, driver mutations are ideal targets for cancer immunotherapies (27). Nevertheless, in most of the tumors, no highly-penetrant mutations have been identified. Furthermore, the mutational landscape of solid tumors is highly heterogeneous, both in terms of cellular composition of a single tumor mass and in terms of individual metastases (28,29). For all these reasons, multiple analyses of multifocal cancers and multiple targeting should be investigated in future trials.
The prognostic and predictive value of the mutational load in the immunotherapy era
The efficacy of immune checkpoint inhibitors indicates that the immune system is actively suppressed in cancer patients, whereas the long-term clinical benefit of only a low percentage of patients might be related to the presence of specific tumor Ags that are recognized by CD8+ and CD4+ T cells in inductive sites, such as lymph nodes or tertiary lymphoid structures in the tumor microenvironment (30), which in turn as effector cells eliminate corresponding tumor cell clones. Although with current technologies, the spatial distribution of anti-tumor T cell specificities in the tumor environment remains uncharacterized, recent data reveals that in different tumor types, the mutanome has a role in shaping the immune landscape, mainly the presence of neoAg-specific CD8+ and CD4+ T cells, either at the level of spontaneous anti-tumor immunity or in the context of conventional cancer therapies (7,10,14). On the other hand, the presence of neoAg-specific CD8+ TILs attracted to the tumor microenvironment by other factors, such as altered expression of cytokines or chemokines, might sculpt the immunogenicity of cancer, a process known as immunoediting (3,31,32).
Several lines of evidence from recent studies support the concept that the mutational and antigenome burden of tumors has a positive prognostic influence. By exploring public RNA-sequencing data, Brown et al. showed that although the number of missense mutations per se was not a prognostic biomarker, the number of immunogenic mutations was associated with increased survival (33). Additionally, the number of immunogenic mutations correlated with higher CD8+ T cell infiltrates, by using as a surrogate the CD8A gene expression, and that this was counterbalanced by increased expression of programmed cell death 1 (PDCD1) and cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) genes, a finding that may have clinical implications (33). In another study, high-dimensional data sets were analyzed in silico and showed an association of neoAg load with a gene signature of cytolytic activity, composed of the combination of the granzyme A (GZMA) and perforin (PRF1) transcripts (34). Importantly, neoAgs, unlike cancer-testis Ags, appear to be depleted during tumor progression maybe due to selective pressure from cytotoxic T cells, a finding supporting the immunoediting hypothesis (34).
Beyond its prognostic influence, recent studies support the hypothesis that the mutational burden has also a positive predictive value in patients treated with immune checkpoint blockade. In a recent study of patients with non-small cell lung carcinoma treated with pembrolizumab (anti-PD-1 blocking monoclonal antibody), an elevated nonsynonymous mutation burden was associated with clinical efficacy that was higher in patients with tumors harboring a smoking signature, i.e., with a higher number of mutations (15). Additionally, the mutational burden correlated with the quantity of immunogenic mutations, which are defined as mutant nonamers that are predicted to have a binding affinity of ≤500 nM to the patient-specific HLA class I alleles.
Along the same line, nonsynonymous mutational load, as well as a high frequency of neoAgs, were associated with clinical benefit in patients with metastatic melanoma treated with ipilimumab (anti-CTLA-4 blocking monoclonal antibody) (35). Moreover, this clinical benefit was associated with a higher expression of the GZMA and PRF1 transcripts that are both upregulated in activated CD8 T cells, as well as with higher expression of immune checkpoint molecules, i.e., CTLA-4 and PD-L2. Importantly, from a clinical point of view, neoAg mutations were not shared between responders. Similar information of a distinct neoAg mutational pattern between tumors was extracted from a large cohort of colorectal carcinoma (CRC) patients (36). In this study, the possible association of antigenicity with immunogenicity was more comprehensively characterized. Hypermutated tumors, i.e., mismatch repair-deficient tumors, but also a group of hypermutated mismatch repair-proficient cases were associated with depletion of regulatory T cells (Tregs)- and myeloid derived suppressor cells (MDSCs)-specific gene expression signatures (metagenes), as well as with upregulation of immune-inhibitory molecules such as CTLA-4, IDO1, PD-1 and PD-L1, whereas in less mutated tumors, i.e., with a mismatch repair-proficient phenotype, either Tregs or the MDSCs metagene signatures where enriched and immune-inhibitory molecules were down-regulated. In line with these findings, the mismatch repair status was a predictor of clinical response with pembrolizumab in a recent small phase II CRC study. Forty percent of patients with mismatch repair-deficient tumors (4 out of 10) showed a partial response to PD-1 blockade, whereas none of the mismatch repair-proficient tumors (0 out of 18) showed any type of responses (37). Remarkably, the average of somatic mutations was 1,782 in mismatch-repair deficient (mean number of neoAgs: 578), as compared with 73 (mean number of neoAgs: 21) in mismatch-repair proficient tumors. Intriguingly, by using an experimental murine sarcoma model, Gubin et al. revealed that following checkpoint blockade immunotherapy, the mutant tumor Ag-specific T cells are targeted and activated and additionally, tumor-specific mutant Ags can be used to create personalized vaccines (25).
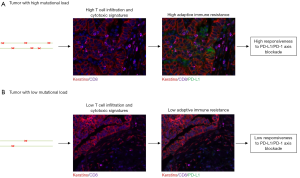
The above data supports the concept that immunotherapeutic interventions can be designed by mapping the mutational and immune landscape of human cancers (38,39). In order to provide stronger evidence that the tumor mutanome can be used as a potential biomarker of clinical benefit in cancer immunotherapies, either alone or in combination with immune landscape signatures, analyses of larger cohorts of patients are required. In these studies, whole-genome or whole-exome sequencing data would be combined with a detailed evaluation of the immune infiltrates, either by using RNA sequencing and gene expression techniques or multiplexed IHC/IF approaches (Figure 2), in order to not only interrogate the spatial distribution of adaptive and innate immunity cell subsets but importantly, to ascribe functional characteristics, as well (40).
Conclusions
The recent surge of publications, focusing on studies investigating the relationship between mutanome and cancer immunity and the development of mutanome-based cancer immunotherapies, many of which already entered the clinic, substantiate the increasing efforts and interests in such a field by both academic institutions and private companies. The circulation of naturally-occurring T lymphocytes specific for Ags derived from mutated cancer genes has been known since the 90’s, thanks to laborious studies of cDNA library screening (41-43). In the same decade, scientists also conceived the first vaccination protocols based on synthetic peptides mimicking the mutant sequences of shared Ags [e.g., RAS (44)]. Since the establishment of the first T cell clones and the discovery of the first immunogenic peptides, more than 20 years ago, the list of cancer-rejection Ags expanded (1), but is probably biased toward a higher proportion of non-mutated shared tumor Ags (e.g., differentiation or over-expressed Ags, etc.), as opposed to shared mutated and cancer-testis Ags. Epitopes generated by processing of hot-spot genes are ideal immunotherapeutic targets because they are highly specific and shared among patients, but recent studies concluded that most of the tumor-specific somatic mutations, estimated to be around 95%, are private and passengers (45). Contemporary scientific and technological breakthroughs convinced onco-immunologists to take into account a different perspective and use individual patient’s HTS data for the identification of immunogenic epitopes and the generation of personalized therapies. In the near future, detailed and enlarged genome databases may supply new shared driver mutations as potential source of therapeutic neoAgs. Indeed, the aforementioned therapeutic approach is still pursued. A clinical trial assessing the safety of a peptide vaccine containing the neoepitope IDH1 is currently ongoing in grade III-IV glioma patients harboring the IDH1R132H-mutation (NCT02454634). Another clinical trial is evaluating a vaccination protocol based on multiple frameshift-derived neoAg-loaded DC of CRC patients with an MSI-positive CRC (NCT01885702).
Recent data suggest that immunotherapy (i.e., ACT and checkpoint inhibitors) exerts a superior clinical effect in patients affected by cancers with a high mutational burden (melanoma, lung, subtypes of colon cancer and bladder) (46). These subjects are important, not only for the identification of predictive and prognostic markers of successful immunotherapies, but also as a source of neoAg-specific T cells, whose nature remains unknown. First, we need to investigate if the absence of central tolerance toward the neoAgs, due to the lack of expression in the thymus, is responsible for a higher functional affinity of the neoAg-specific T cells, ultimately mediating a superior cytotoxic activity and tumor regression. Alternatively, T cells may display peculiar properties, such as high proliferative capacity and pluripotency (i.e., non-terminally differentiated cells). Moreover, other key aspects that deserve more in-depth studies are the role of neoAg-specific CD4+ T cells that have been shown to represent the main component of anti-neoAg reactivities in some tumors (9,10,23), the role of the tumor microenvironment and the relative contribution of phenomena of tumor editing and Ag spreading. This knowledge is critical to fully exploit neoAg-specific T cell potentiality and to better define the immunotherapeutic strategies. It is conceivable to think about future trials combining different therapeutic approaches, including conventional therapies. For example, randomized phase II clinical trials are exploring combinations of DC vaccines with radiation therapy and pre-vaccination IFN-α (NCT01973322).
Emerging data suggests that neoAg-specific T cells can also be detected in tumors characterized by a low mutational burden (23). Nevertheless, these patients could benefit from alternative active immunotherapeutic strategies, not requiring the identification of neoAgs, such as whole tumor vaccines. Indeed, autologous tumor vaccines provide a source for the full repertoire of the patient-specific TAAs, including its private neoAgs. This personalized approach may be critical in case of poor tumor immunogenicity and low mutational load in our efforts to induce or boost the adaptive immunity, to in vivo select the Ags with the higher therapeutic potential and to create the optimal milieu for further therapeutic interventions (e.g., vaccination and/or ACT).
Acknowledgements
PG Foukas is supported by a grant from Fondation Emma Muschamp.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Coulie PG, Van den Eynde BJ, et al. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer 2014;14:135-46. [Crossref] [PubMed]
- Castle JC, Kreiter S, Diekmann J, et al. Exploiting the mutanome for tumor vaccination. Cancer Res 2012;72:1081-91. [Crossref] [PubMed]
- Matsushita H, Vesely MD, Koboldt DC, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 2012;48:400-4. [Crossref] [PubMed]
- Yadav M, Jhunjhunwala S, Phung QT, et al. Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 2014;515:572-6. [Crossref] [PubMed]
- Robbins PF, Lu YC, El-Gamil M, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med 2013;19:747-52. [Crossref] [PubMed]
- van Rooij N, van Buuren MM, Philips D, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol 2013;31:e439-42. [Crossref] [PubMed]
- Wick DA, Webb JR, Nielsen JS, et al. Surveillance of the tumor mutanome by T cells during progression from primary to recurrent ovarian cancer. Clin Cancer Res 2014;20:1125-34. [Crossref] [PubMed]
- Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2014;371:2189-99. [Crossref] [PubMed]
- Kreiter S, Vormehr M, van de Roemer N, et al. Mutant MHC class II epitopes drive therapeutic immune responses to cancer. Nature 2015;520:692-6. [Crossref] [PubMed]
- Linnemann C, van Buuren MM, Bies L, et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat Med 2015;21:81-5. [Crossref] [PubMed]
- Tran E, Turcotte S, Gros A, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science 2014;344:641-5. [Crossref] [PubMed]
- Lennerz V, Fatho M, Gentilini C, et al. The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc Natl Acad Sci U S A. 2005;102:16013-8. [Crossref] [PubMed]
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013;339:1546-58. [Crossref] [PubMed]
- Rajasagi M, Shukla SA, Fritsch EF, et al. Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood 2014;124:453-62. [Crossref] [PubMed]
- Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124-8. [Crossref] [PubMed]
- Kreiter S, Castle JC, Türeci O, et al. Targeting the tumor mutanome for personalized vaccination therapy. Oncoimmunology 2012;1:768-9. [Crossref] [PubMed]
- Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415-21. [Crossref] [PubMed]
- Lu YC, Robbins PF. Cancer immunotherapy targeting neoantigens. Semin Immunol 2016;28:22-7. [Crossref] [PubMed]
- Desrichard A, Snyder A, Chan TA. Cancer Neoantigens and Applications for Immunotherapy. Clin Cancer Res 2016;22:807-12. [Crossref] [PubMed]
- Vormehr M, Diken M, Boegel S, et al. Mutanome directed cancer immunotherapy. Curr Opin Immunol 2016;39:14-22. [Crossref] [PubMed]
- Lundegaard C, Lund O, Nielsen M. Prediction of epitopes using neural network based methods. J Immunol Methods 2011;374:26-34. [Crossref] [PubMed]
- Nielsen M, Lund O, Buus S, et al. MHC class II epitope predictive algorithms. Immunology 2010;130:319-28. [Crossref] [PubMed]
- Tran E, Ahmadzadeh M, Lu YC, et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 2015;350:1387-90. [Crossref] [PubMed]
- Andersen RS, Thrue CA, Junker N, et al. Dissection of T-cell antigen specificity in human melanoma. Cancer Res 2012;72:1642-50. [Crossref] [PubMed]
- Gubin MM, Zhang X, Schuster H, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 2014;515:577-81. [Crossref] [PubMed]
- Cohen CJ, Gartner JJ, Horovitz-Fried M, et al. Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J Clin Invest 2015;125:3981-91. [Crossref] [PubMed]
- Martincorena I, Roshan A, Gerstung M, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 2015;348:880-6. [Crossref] [PubMed]
- Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883-92. [Crossref] [PubMed]
- Yachida S, Jones S, Bozic I, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 2010;467:1114-7. [Crossref] [PubMed]
- Dieu-Nosjean MC, Goc J, Giraldo NA, et al. Tertiary lymphoid structures in cancer and beyond. Trends Immunol 2014;35:571-80. [Crossref] [PubMed]
- DuPage M, Mazumdar C, Schmidt LM, et al. Expression of tumour-specific antigens underlies cancer immunoediting. Nature 2012;482:405-9. [Crossref] [PubMed]
- Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science 2011;331:1565-70. [Crossref] [PubMed]
- Brown SD, Warren RL, Gibb EA, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res 2014;24:743-50. [Crossref] [PubMed]
- Rooney MS, Shukla SA, Wu CJ, et al. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015;160:48-61. [Crossref] [PubMed]
- Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207-11. [Crossref] [PubMed]
- Angelova M, Charoentong P, Hackl H, et al. Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol 2015;16:64. [Crossref] [PubMed]
- Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 2015;372:2509-20. [Crossref] [PubMed]
- Martin SD, Coukos G, Holt RA, et al. Targeting the undruggable: immunotherapy meets personalized oncology in the genomic era. Ann Oncol 2015;26:2367-74. [PubMed]
- Becht E, Giraldo NA, Germain C, et al. Immune Contexture, Immunoscore, and Malignant Cell Molecular Subgroups for Prognostic and Theranostic Classifications of Cancers. Adv Immunol 2016;130:95-190. [Crossref] [PubMed]
- Yuan J, Hegde PS, Clynes R, et al. Novel technologies and emerging biomarkers for personalized cancer immunotherapy. J Immunother Cancer 2016;4:3. [Crossref] [PubMed]
- Sibille C, Chomez P, Wildmann C, et al. Structure of the gene of tum- transplantation antigen P198: a point mutation generates a new antigenic peptide. J Exp Med 1990;172:35-45. [Crossref] [PubMed]
- Wölfel T, Hauer M, Schneider J, et al. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science 1995;269:1281-4. [Crossref] [PubMed]
- Wang RF, Wang X, Atwood AC, et al. Cloning genes encoding MHC class II-restricted antigens: mutated CDC27 as a tumor antigen. Science 1999;284:1351-4. [Crossref] [PubMed]
- Gjertsen MK, Bakka A, Breivik J, et al. Vaccination with mutant ras peptides and induction of T-cell responsiveness in pancreatic carcinoma patients carrying the corresponding RAS mutation. Lancet 1995;346:1399-400. [Crossref] [PubMed]
- Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science 2011;331:1553-8. [Crossref] [PubMed]
- Gubin MM, Schreiber RD. CANCER. The odds of immunotherapy success. Science 2015;350:158-9. [Crossref] [PubMed]


