
The silencing of lnc-NONHSAT071210 suppresses the proliferation, fibrosis, migration, and invasion of TGFβ1-treated lung epithelial cells
Highlight box
Key findings
• The role of lnc-NONHSAT071210 in CTD-associated pulmonary fibrosis.
What is known and what is new?
• lnc-NONHSAT071210 depletion decreased the proliferation and fibrosis in the TGFβ1-treated lung epithelial cells.
• lnc-NONHSAT071210 might be a potential biomarker for CTD-associated pulmonary fibrosis.
What is the implication, and what should change now?
• The findings provide new insights into the pathogenesis and targeted therapy of CTD-associated pulmonary fibrosis.
Introduction
Connective tissue disease (CTD) is a heterogeneous autoimmune disease characterized by an inflammatory response in the blood vessels and connective tissues (1). Due to the abundant connective tissues in the bronchus, pulmonary vessels, lung interstitium, pleura, the lung is one of the organs most susceptible to CTD (2). The accumulation of immune complexes in the lung tissues mediated by CTD may induce the diffuse infiltration of inflammatory cells, result in the massive deposit of interstitial cells and extracellular matrix (ECM) components in interstitial tissues, and even damage pulmonary alveoli and blood capillaries (3). Consequently, the iterative repair and damage of pulmonary interstitial tissues may contribute to pulmonary modeling and eventually trigger pulmonary fibrosis (3). Pulmonary fibrosis is a common type of CTD-associated interstitial lung disease (CTD-ILD), and is a severe complication of CTD (4). Progressive fibrosis in the lungs may cause a loss of lung function, which is a leading cause in morbidity and mortality (5,6). Despite deeper understandings of the etiology of CTD-associated pulmonary fibrosis, its treatment remains a great challenge (7).
Long non-coding ribonucleic acids (lncRNAs) refer to a class of transcripts, over 200 nts in length, that are unable to code proteins (8). Numerous studies have sought to identify lncRNAs, as these are promising biomarkers for human malignancies and diseases (9,10). Several lncRNAs have been shown to have important functions in the pathogenesis of systemic sclerosis, dermatomyositis, and cutaneous lupus erythematosus (11). Notably, lncRNAs have been shown to participate in pulmonary fibrosis (12-14). Lnc-NONHSAT071210 is a novel lncRNA on which little research has been conducted to date.
This study sought to examine the expression of lnc-NONHSAT071210 in CTD-associated pulmonary fibrosis patients and its effects on the biological behaviors in a transforming growth factor β1 (TGFβ1)–induced pulmonary fibrosis model. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5223/rc).
Methods
Microarray data
Experimental sets from the Affymetrix Gene Chip Human Transcriptome Array 2.0 were used to identify the differentially expressed lncRNAs in CTD-associated pulmonary fibrosis, and the expression of the top 7 most significantly upregulated lncRNAs was displayed in heatmaps.
Clinical samples
CTD-associated pulmonary fibrosis tissues and normal lung tissues were obtained from Liuzhou People’s Hospital. Patients with overlapping syndromes or multiple autoimmune diseases were excluded from the study. The lung samples obtained from the CTD-associated pulmonary fibrosis patients undergoing lung transplantation were stored at −80 ℃. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Liuzhou People’s Hospital and informed consent was taken from all the patients.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was prepared from the samples or cells using TRIzol®, and the RNA was converted to complementary deoxyribonucleic acid (DNA) (Sangon Biotech Co., Ltd.) using the Omniscript® RT Kit (Qiagen). The RT-qPCR was performed using the Synergy Brands (SYBR)-Green dye method (Clontech Laboratories, Inc.) in accordance with the manufacturer’s instructions. The thermocycling conditions were as follows: denaturation at 95 ℃ for 5 min, followed by 40 cycles of 95 ℃ for 10 sec and 60 ℃ for 30 sec. The relative messenger RNA levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and calculated using the 2–∆∆Cq method.
Cell culture and treatment
F-12K medium (Wisent Biotechnology) supplemented with fetal bovine serum (FBS) was used to culture the human lung epithelial cell line A549, which was obtained from the American Type Culture Collection. The cells were incubated in a humidified atmosphere containing 5% carbon dioxide at 37 ℃. The recombinant human TGFβ1 (Procell Life Science and Technology Co., Ltd.) was used to treat the A549 cells to mimic pulmonary fibrosis injury in vitro.
Plasmid transfection
Lentivirus-mediated short-hairpin RNAs (shRNAs) targeting lnc-NONHSAT071210 (shRNA-071210-#1#2#3) or its negative control (shRNA-NC) were obtained from TsingKe Inc. Plasmid transfection was performed with the Lipofectamine 2000 (GE Healthcare Life Sciences) before the TGF-β1 treatment. The cells were collected 48 h later for the subsequent experiments.
Cell Counting Kit-8 (CCK-8)
In accordance with the product manual in the CCK-8 kit (Wanlei Bio, Co., Ltd.), cell viability was examined. In brief, the transfected cells were seeded in a 96-well plate at 37 ℃ and exposed to TGFβ1. CCK-8 solution (10 µL) was supplemented for an extra 2 h, and the OD 450 nm value was monitored by a microplate reader (Eppendorf, Hamburg, Germany).
Flow cytometry analysis
The transfected cell suspension exposed to TGFβ1 underwent fixation with 70% ethanol at 4 ℃ overnight. Next, the cells were stained with a solution containing propidium iodide (PI; Shanghai Yeasen Biotechnology Co., Ltd.), and 100 µL of Ribonuclease A (RNase A) for 30 min at 37 ℃. Finally, a flow cytometer (Partec, Arlesheim, Switzerland) was used to analyze cell cycle distribution.
ELISAs
Following the treatment with TGFβ1, the transfected A549 cells were inoculated in 6-well plates (2×105 cells/mL), and the supernatants were harvested. Type I collagen (COL1) levels were examined in accordance with the manufacturer’s instructions by corresponding enzyme-linked immunosorbent assay (ELISA) kits obtained from Nanjing Jiancheng Bioengineering Institute. A microplate reader (Eppendorf, Hamburg, Germany) was used to examine OD 450 nm values.
Wound-healing assays
The monolayers of the A549 cells inoculated in the 6-well plates (4×105 cells/well) were scraped using a 200-µL pipette tip when 80% confluency was reached. The suspended cells were then submerged in serum-starved F-12K medium. A light microscope (Olympus Corporation) was used to document the wound area at 0 h, 24 h, and 48 h.
Transwell invasion assays
The upper sides of Matrigel-coated transwell inserts (Beijing Unique Biotech Co., Ltd.) were loaded with A549 cells (2×105/mL) in serum-starved medium, and the undersides were supplemented with 600 µL F-12K medium comprising 10% FBS. The invaded cells were dyed with crystal violet (Aladdin, China) 24 h later, and were counted using a fluorescence microscope (Leica DM3000k).
Western blot
The prepared protein samples were quantified using the bicinchoninic acid (BCA) protein assay kit (CWBIO Corporation) and protein extraction kit (Beijing ComWin Biotech Co., Ltd.). Next, Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE) was used to divide and transfer the protein samples to the Polyvinylidene Fluoride (PVDF) membranes. The cells were incubated in 5% skim milk for 1 hour to block non-specific binding. Subsequently, the membranes were probed with primary antibodies at 4 ℃ overnight and with horseradish peroxidase–linked anti-rabbit (cat. no. ab6721; 1:2000; Abcam) secondary antibody on the following day. The enhanced chemiluminescence detection system was used to visualize the protein bands, and band intensity was determined using LI-COR Image Studio v5.2.5 Software (LI-COR Biosciences). The value was normalized to GAPDH. Anti-TGFβ1 (cat. No. 21898-1-AP; 1:1,000; Proteintech), anti- alpha smooth muscle actin (α-SMA; cat. No. #19245; 1:1,000; Cell Signaling Technology), and anti-GAPDH (cat. No. ab9485; 1:2,500; Abcam) were used.
Statistical analyses
All the statistical analyses were conducted using GraphPad Prism 8 software (GraphPad Software, Inc.), and the continuous variables are presented as the mean ± standard deviation (SD) from 3 independent experiments. A 1-way analysis of variance followed by Tukey’s test was used for comparisons among multiple groups. The Student’s t test was used for comparisons between 2 groups. A P value <0.05 was considered statistically significant.
Results
Lnc-NONHSAT071210 expression was increased in the tissues of the CTD-associated pulmonary fibrosis patients
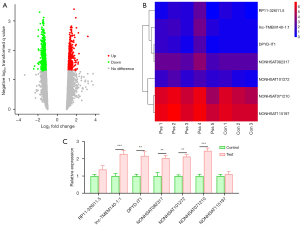
To identify the genes associated with CTD-associated pulmonary fibrosis, the Affymetrix Human GeneChip was applied and the upregulation or downregulation of the genes was recorded. The data revealed that 1688 lncRNAs were differentially expressed between the CTD-associated pulmonary fibrosis tissues and normal tissues (Figure 1A). Subsequently, the top 7 most significantly upregulated lncRNAs (i.e., RP11-326I11.5, lnc-TMEM140-1:1, DPYD-IT1, NONHSAT082317, NONHSAT101272, NONHSAT071210, and NONHSAT115197) were selected, the expression of which was depicted in a heatmap (Figure 1B). As Figure 1C shows, the RT-qPCR analysis revealed that compared to the control group, lnc-NONHSAT071210 was the most highly overexpressed lncNRA in the CTD-associated pulmonary fibrosis tissues among the 7 selected lncRNAs (Figure 1C). Thus, the expression of Lnc-NONHSAT071210 was increased in the tissues of the CTD-associated pulmonary fibrosis patients.
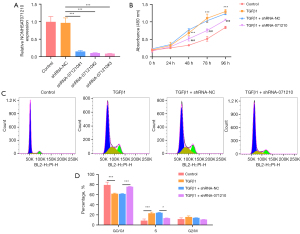
Interference with lnc-NONHSAT071210 decreases the proliferation and cell cycle in TGFβ1-treated A549 cells
To examine the role of lnc-NONHSAT071210 in CTD-associated pulmonary fibrosis, lnc-NONHSAT071210 was silenced via the transduction of shRNA-071210#1#2#3 plasmids, and the efficacy was verified by RT-qPCR. Notably, shRNA-071210#3 had a better interference efficacy and thus was chosen for the subsequent experiments (Figure 2A). The CCK-8 assays showed that TGFβ1 treatment significantly increased the viability of the A549 cells and the silencing of lnc-NONHSAT071210 significantly decreased the viability of the TGFβ1-challenged A549 cells (Figure 2B). The results of the flow cytometry analysis showed that TGFβ1 exposure decreased the G0/G1 ratio and increased the S phase. Additionally, the shortage of lnc-NONHSAT071210 increased the G0/G1 ratio and decreased the ratio of the S phase (Figure 2C,2D). Thus, lnc-NONHSAT071210 interference inhibited TGFβ1-induced proliferation and the cell cycle of the A549 cells.
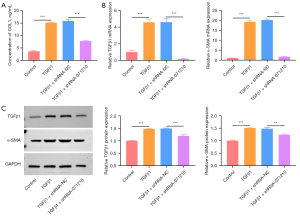
Knockdown of lnc-NONHSAT071210 decreases TGFβ1-induced fibrosis in A549 cells
The level of COL1, a characteristic of fibrosis, was increased in the TGFβ1-treated A549 cells, and the deficiency of lnc-NONHSAT071210 decreased the CoL1 level in the TGFβ1-treated A549 cells (Figure 3A). The RT-qPCR and western blot analysis showed that the increased TGFβ1 and α-SMA expression in the TGFβ1-challenged A549 cells was significantly reduced after lnc-NONHSAT071210 had been knocked down (Figure 3B,3C). Thus, lnc-NONHSAT071210 downregulation significantly decreased TGFβ1-stimulated fibrosis in the A549 cells.
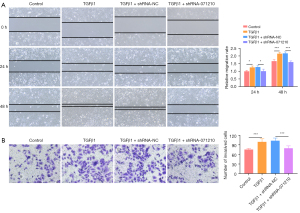
Lnc-NONHSAT071210 deficiency suppresses TGFβ1-induced migration and invasion of A549 cells
As Figure 4A,4B show, the supplementation of TGFβ1 significantly increased the migratory and invasive abilities of A549 cells, while the inhibition of lnc-NONHSAT071210 significantly decreased the migratory and invasive abilities of TGFβ1-challenged A549 cells. Thus, lnc-NONHSAT071210 protected against fibronectin-driven A549 cell migration and invasion.
Discussion
CTD-ILD is the most serious complication of CTD, which has high disability and mortality rates (4). The study has shown that the main pathogenesis of CTD-associated pulmonary fibrosis is the continuous proliferation of alveolar epithelial type II cells and fibroblasts, which results in collagen deposition and further leads to the irreversible disease process (15). TGFβ is a pleiotropic cytokine that has been implicated in multiple biological processes, including inflammation, immunoregulation, hematopoiesis, tissue remodeling, and tumor development (16-18). Research has highlighted the important role of TGFβ, which has been shown to stimulate the production of ECM components as represented by Collagen I and fibronectin, in organ fibrosis (19). This study used TGFβ1 to treat the human lung epithelial cell line A549 to construct a pulmonary fibrosis model in vitro and then examined cell proliferation, cell cycle, fibrosis, migration, and invasion to determine the progression of CTD-associated pulmonary fibrosis.
LncRNAs have been confirmed to be engaged in a variety of cellular events and human diseases through the regulation of protein-coding genes at the transcriptional and post-transcriptional levels (20). In recent years, extensive evidence has been gathered showing the significance of lncRNAs in CTD (11,21). Notably, the abnormal expression of lncRNAs may affect the process of pulmonary fibrosis and lncRNAs may be valuable biomarkers for pulmonary fibrosis. For example, lncRNA Hoxaas3 accelerates pulmonary fibrosis by targeting the miR-450b-5p/Runx1 axis (22). Liu et al. found that lncRNA SNHG16 also functions as a promoter in pulmonary fibrosis via the modulation of the Notch2 pathway (12). Sun et al. showed that lncRNA pulmonary fibrosis inhibitor (PFI) ameliorates pulmonary fibrosis by binding to splicing regulator Serine/arginine-rich splicing factor 1 (SRSF1) (14). Lnc-NONHSAT071210 is a novel lncRNA that has not been examined previously. Using the Affymetrix Human GeneChip, this study showed that lnc-NONHSAT071210 was significantly overexpressed in the tissues of CTD-associated pulmonary fibrosis patients and this was further confirmed by RT-qPCR.
Based on our findings, we hypothesized that lnc-NONHSAT071210 is related to the development of CTD-associated pulmonary fibrosis. For further validation, lnc-NONHSAT071210 was knocked down to evaluate the effects of lnc-NONHSAT071210 silencing on the proliferation, cell cycle, fibrosis, migration, and invasion in the TGFβ1-challenged A549 cells. The results of the functional experiments showed that TGFβ1 treatment significantly enhanced the viability and cell cycle of the A549 cells, and lnc-NONHSAT071210 deficiency significantly impeded the viability and cell cycle of the TGFβ1-challenged A549 cells. In addition, it is well documented that TGFβ1 is a potent pro-fibrogenic cytokine (23). α-SMA is a marker for myofibroblasts responsible for the contractility of fibroblasts (24). CoL1, which is secreted by myofibroblasts that elicit critical activities during the process of fibrosis, is a major component of the ECM (25). We found that the TGFβ1 treatment significantly increased TGFβ1 and α-SMA expression and increased edCoL1 content. Following interference with lnc-NONHSAT071210, the increased TGFβ1 and α-SMA expression and CoL1 content were both decreased. Thus, lnc-NONHSAT071210 appears to mitigate TGFβ1-induced fibrosis in A549 cells. Further, our experimental results also revealed that the increased migratory and invasive abilities of the TGFβ1-insulted A549 cells were both increased by silencing lnc-NONHSAT071210.
Collectively, lnc-NONHSAT071210 depletion decreased the proliferation, cell cycle, fibrosis, migration, and invasion in the TGFβ1-treated A549 cells. Thus, lnc-NONHSAT071210 might be a potential biomarker for CTD-associated pulmonary fibrosis. This is the first study to reveal the role of lnc-NONHSAT071210 in CTD-associated pulmonary fibrosis. Our findings provide novel insights into the pathogenesis of CTD-associated pulmonary fibrosis and the exploration of targeted therapies for CTD-associated pulmonary fibrosis. Nonetheless, there are still some challenges in the therapies based on lncRNAs. For instance, the structural and functional information of most lncRNAs remains uncharacterized in human diseases. Moreover, lncRNAs are poorly conserved across species. Hence, it is of great necessity to identify valuable lncRNAs in specific human diseases and explore novel lncRNAs. In the future, animal studies need to be conducted to further verify the role of lnc-NONHSAT071210 in CTD-associated pulmonary fibrosis. Additionally, the downstream mechanism of lnc-NONHSAT071210 requires exploration. Finally, consideration also needs to be given to whether any signaling pathways are involved in lnc-NONHSAT071210-mediated CTD-associated pulmonary fibrosis.
Conclusions
The role of lnc-NONHSAT071210 in CTD-associated pulmonary fibrosis was investigated. It revealed that lnc-NONHSAT071210 may be a potential biomarker for CTD-associated pulmonary fibrosis. The findings provide new insights into the pathogenesis and targeted therapy of CTD-associated pulmonary fibrosis.
Acknowledgments
Funding: This study was supported by National Natural Science Foundation of China (No. 81960304) and Liuzhou Scientific and Technological Projects (No. 2020NBAA0804).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5223/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5223/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5223/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Liuzhou People’s Hospital and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lefeuvre L, Dan D. Connective tissue disease-related interstitial lung disease. Rev Med Suisse 2021;17:492-6. [Crossref] [PubMed]
- Shao T, Shi X, Yang S, et al. Interstitial Lung Disease in Connective Tissue Disease: A Common Lesion With Heterogeneous Mechanisms and Treatment Considerations. Front Immunol 2021;12:684699. [Crossref] [PubMed]
- Saketkoo LA, Mittoo S, Huscher D, et al. Connective tissue disease related interstitial lung diseases and idiopathic pulmonary fibrosis: provisional core sets of domains and instruments for use in clinical trials. Thorax 2014;69:428-36. [Crossref] [PubMed]
- Luppi F, Sebastiani M, Salvarani C, et al. Acute exacerbation of interstitial lung disease associated with rheumatic disease. Nat Rev Rheumatol 2022;18:85-96. [Crossref] [PubMed]
- Spagnolo P, Distler O, Ryerson CJ, et al. Mechanisms of progressive fibrosis in connective tissue disease (CTD)-associated interstitial lung diseases (ILDs). Ann Rheum Dis 2021;80:143-50. [Crossref] [PubMed]
- Noble PW, Barkauskas CE, Jiang D. Pulmonary fibrosis: patterns and perpetrators. J Clin Invest 2012;122:2756-62. [Crossref] [PubMed]
- Wallace B, Vummidi D, Khanna D. Management of connective tissue diseases associated interstitial lung disease: a review of the published literature. Curr Opin Rheumatol 2016;28:236-45. [Crossref] [PubMed]
- Jarroux J, Morillon A, Pinskaya M. History, Discovery, and Classification of lncRNAs. Adv Exp Med Biol 2017;1008:1-46. [Crossref] [PubMed]
- Chi Y, Wang D, Wang J, et al. Long Non-Coding RNA in the Pathogenesis of Cancers. Cells 2019;8:1015. [Crossref] [PubMed]
- Zhang X, Hong R, Chen W, et al. The role of long noncoding RNA in major human disease. Bioorg Chem 2019;92:103214. [Crossref] [PubMed]
- Muntyanu A, Le M, Ridha Z, et al. Novel role of long non-coding RNAs in autoimmune cutaneous disease. J Cell Commun Signal 2021; Epub ahead of print. [Crossref] [PubMed]
- Liu P, Zhao L, Gu Y, et al. LncRNA SNHG16 promotes pulmonary fibrosis by targeting miR-455-3p to regulate the Notch2 pathway. Respir Res 2021;22:44. [Crossref] [PubMed]
- Xu Q, Cheng D, Liu Y, et al. LncRNA-ATB regulates epithelial-mesenchymal transition progression in pulmonary fibrosis via sponging miR-29b-2-5p and miR-34c-3p. J Cell Mol Med 2021;25:7294-306. [Crossref] [PubMed]
- Sun J, Jin T, Su W, et al. The long non-coding RNA PFI protects against pulmonary fibrosis by interacting with splicing regulator SRSF1. Cell Death Differ 2021;28:2916-30. [Crossref] [PubMed]
- Montero P, Milara J, Roger I, et al. Role of JAK/STAT in Interstitial Lung Diseases; Molecular and Cellular Mechanisms. Int J Mol Sci 2021;22:6211. [Crossref] [PubMed]
- Travis MA, Sheppard D. TGF-β activation and function in immunity. Annu Rev Immunol 2014;32:51-82. [Crossref] [PubMed]
- Liu S, Ren J, Ten Dijke P. Targeting TGFβ signal transduction for cancer therapy. Signal Transduct Target Ther 2021;6:8. [Crossref] [PubMed]
- Chen W, Ten Dijke P. Immunoregulation by members of the TGFβ superfamily. Nat Rev Immunol 2016;16:723-40. [Crossref] [PubMed]
- Meng XM, Nikolic-Paterson DJ, Lan HY. TGF-β: the master regulator of fibrosis. Nat Rev Nephrol 2016;12:325-38. [Crossref] [PubMed]
- Dykes IM, Emanueli C. Transcriptional and Post-transcriptional Gene Regulation by Long Non-coding RNA. Genomics Proteomics Bioinformatics 2017;15:177-86. [Crossref] [PubMed]
- Cai B, Cai J, Yin Z, et al. Long non-coding RNA expression profiles in neutrophils revealed potential biomarker for prediction of renal involvement in SLE patients. Rheumatology (Oxford) 2021;60:1734-46. [Crossref] [PubMed]
- Lin S, Zhang R, Xu L, et al. LncRNA Hoxaas3 promotes lung fibroblast activation and fibrosis by targeting miR-450b-5p to regulate Runx1. Cell Death Dis 2020;11:706. [Crossref] [PubMed]
- Ihn H. Pathogenesis of fibrosis: role of TGF-beta and CTGF. Curr Opin Rheumatol 2002;14:681-5. [Crossref] [PubMed]
- Shinde AV, Humeres C, Frangogiannis NG. The role of α-smooth muscle actin in fibroblast-mediated matrix contraction and remodeling. Biochim Biophys Acta Mol Basis Dis 2017;1863:298-309. [Crossref] [PubMed]
- Seo BR, Chen X, Ling L, et al. Collagen microarchitecture mechanically controls myofibroblast differentiation. Proc Natl Acad Sci U S A 2020;117:11387-98. [Crossref] [PubMed]
(English Language Editor: L. Huleatt)


