
Investigation of hub genes and immune infiltration in androgenetic alopecia using bioinformatics analysis
Introduction
Androgenetic alopecia (AGA), also known as androgenic alopecia (1), is a type of non-scarring hair loss mainly characterized by a receding hairline on the forehead and sideburns, while the temporal and occipital areas remain unaffected, forming a characteristic “horseshoe” pattern (2), which is a intricated disorder with the interplay of genetic, mental, and environmental factors precipitating its onset (3). Intriguingly, the clinical manifestation of AGA is not identical in males and females, and the latter presents as top hair thinning and diffuse hair loss or a Christmas tree pattern (4). The prevalence of AGA varies by ethnicity and age (5), with a higher incidence in males than females (6). Despite taking a benign clinical course, the negative influence on the life quality of AGA patients is profoundly substantial (7), especially in psychological terms; it can be associated with depression, anxiety, and psychological illnesses (1,8), taking a considerable toll on individuals and societies. Currently, finasteride and minoxidil are the only two types of drugs approved by the US Food and Drug Administration (FDA) (9) for the management of AGA; however, certain drug reactions and a high recurrence rate limit the value of these drugs (10). Besides, despite the straightforward diagnostic approach of AGA in clinical settings, its characteristics of hair loss are sometimes similar to other types of disorders (e.g., telogen effluvium, alopecia areata) (11,12). Thus, diagnostic biomarkers and therapeutic targets for AGA remain in demand.
Pathologically, the hair follicle (HF) takes the center stage in the initiation and progression of AGA, which manifests as shortening of the anagen phase and prolongation of the telogen phase, eventually contributing to miniaturization and progressive hair loss (13,14). Notably, HF has its own unique immune system (15), which ensures normal hair growth and plays a pivotal role in immune protection (16,17), with compelling evidence of in vivo data elucidating that immune cells function as an activator for hair follicle stem cells (HFSCs) and the telogen-to-anagen transition of HF (16,18,19). Besides the immune component, HFSCs are also essential parts of the local environment that participate in the cycle of hair regeneration which remain quiescent and fail to differentiate into HF precursors in the alopecia region of AGA (20). Previous research elucidated the intricate and dynamic interplay among various components of the local environment of HF, thus affording an opportunity to overcome the lack of major breakthroughs against AGA.
In the work presented here, the GSE90594 dataset, which contains scalp skin vertex biopsies from 14 cases of male AGA and healthy volunteers, was used to investigate the expression patterns of AGA, thereby identifying the differentially expressed genes (DEGs). Functional enrichment analysis was subsequently performed to reveal the biological functions and related pathways of the DEGs. Next, a protein-protein interaction (PPI) network was constructed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database and the cytoHubba plugin of Cytoscape (https://cytoscape.org/) was used to score each node gene with 10 algorithms, then the intersection analysis was performed to obtain the key genes via R package “UpSet. Thereafter, the Cell-type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT) algorithm was performed to evaluate the relative abundance of immune cell between male AGA patients and healthy controls, the correlation analysis between key genes and infiltrating immune cells was also analyzed, and the significant immune-cell related genes (IRGs) were retained and intersected with the DEGs between immortalized balding and non-balding human dermal papilla cells (DPCs) in the GSE93766 dataset as well as the DEGs obtained by GSE90594 dataset, thus obtaining the hub genes of AGA. Finally, the hub genes were validated using the GSE36169, which contained expression profiling of biopsied from haired and bald scalp of five individuals with AGA. Collectively, we could discover that the hub genes identified in this study are closely linked with the causative factors of AGA, which could be used as the viable diagnostic and therapeutic target in the clinical applications. We present the following article in accordance with the STREGA reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4634/rc).
Methods
Raw data acquisition
Transcriptomic expression profiling of AGA (GSE90594) was downloaded from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo) database. The dataset was based on GPL17077 platform, including scalp skin vertex biopsies from 14 cases of male AGA and a matched numbers of healthy volunteers. Besides, GSE36169 was also downloaded, which contained expression profiling of biopsied from haired and bald scalp of five individuals with AGA, for the analysis. Thereafter, high throughput sequencing data of GSE93766 based on GPL11154 platform was also acquired from GEO database, containing immortalized balding and non-balding human DPCs derived from male AGA patients. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Data preprocessing
After expression matrixes of GSE90594 and GSE36169 were downloaded from the Gene Expression Omnibus (GEO) database, a platform annotation file was downloaded to transfer the probe expression matrix into a gene expression matrix. Then, we downloaded all SRA files of the GSE93766 dataset, including SRR5184331, SRR5184332, SRR5184333, SRR5184340, SRR5184341, and SRR5184342. Next, the fasterq-dump of R package “sratoolkit” was used to transfer the SRA files to the fastq files, and “fastqc” and “multiqc” were used to examine the quality of sequencing. Thereafter, package “Trim Galore” were used to proceed automated adapter and quality trimming, and the gene expression matrixes were obtained using the featureCounts function of “Rsubread” package.
Principal component analysis
Principal component analysis (PCA) of samples in the GSE90594 dataset was analyzed by R package “FactoMineR”, and visualized by R package “factoextra”.
Identifications of DEGs
The DEGs in datasets used were identified using R package “DESeq2”, with the P value <0.05 and |log fold change (logFC)| >0.5 as the threshold, then visualized by R package “ggplot2” and package “pheatmap”.
Functional enrichment analysis of DEGs
The R package “org.Hs.eg.db” was used to transform gene names of DEGs into gene ID. Next, R package “ClusterProfiler” was used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses, with the P value <0.05 as selection criteria, thus visualizing by package “GOplot” and package “ggplot2”.
Screening of key genes of DEGs
First, the STRING (https://string-db.org) database was conducted to construct the PPI network of DEGs with minimum required interaction score of 0.4, thereby obtaining the interaction network file. Next, the cytoHubba plugin of Cytoscape (v 3.8.2) was used to score each node gene with the 10 algorithms, containing MCC (Maximal Clique Centrality), DMNC (Density of Maximum Neighborhood Component), MNC (Maximum Neighborhood Component), Degree, EPC (Edge Percolated Component), BottleNeck, EcCentricity, Closeness, Radiality, and Betweenness. After the top 50 node genes ranked by each algorithm were obtained, the intersection analysis was performed to obtain the key genes via R package “UpSet”.
Immune infiltration and correlation analysis
The CIBERSORT algorithm was performed to evaluate the relative abundance of immune cell between male AGA patients and healthy controls. First, the expression matrix data of GSE90594 were standardized via package “DESeq2”. Then, based on the reference gene expression signatures of 22 kinds of immune cell gene set provided by Newman et al. (21), the expression matrix of GSE90594 was used for calculating the relative abundance of infiltrating immune cells with 1,000 permutations (22). Specifically, only results with P<0.05 were retained for the following analyses, and the visualization of results obtained from CIBERSORT was performed using packages “vioplot”, “pheatmap”, and “ggplot2”. Besides, the correlation analysis between key genes and infiltrating immune cells was also analyzed, and the significant IRGs were retained and intersected with the DEGs between immortalized balding and non-balding human DPCs in the GSE93766 dataset, thus obtaining the hub genes of AGA.
Statistical analysis
Correlation between key genes and relative levels of immune cell subtypes were carried out through Spearman’s correlation analysis, and comparisons of expression levels of each hub genes between male AGA patients and healthy controls were performed using Wilcoxon test. A P value <0.05 was considered statistically significant in the above-mentioned analyses.
Results
Identification of DEGs
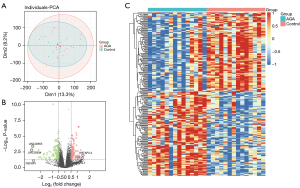
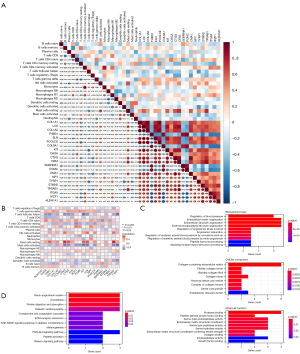
To investigate the expression patterns of AGA, the GSE90594 dataset was used for the following analyses, containing scalp skin vertex biopsies from 14 cases of male AGA patients and an identical number of healthy volunteers. Notably, the results of PCA manifested that the differences between AGA patients and healthy controls are limited (Figure 1A). Next, we further obtained the DEGs between different samples with P<0.05 and |logFC)| >0.5, thus identifying a total of 234 DEGs, which included 114 up-regulated and 120 down-regulated genes, and visualized them with a volcano plot (Figure 1B) and heatmap (Figure 1C), among which LINC02655, LINC02634, and PSPHP1 were the DEGs with the most significant logFC value, whose expression levels were consistently diminished in the AGA samples. Collectively, the above-mentioned results indicated that the expression pattern between AGA patients and healthy samples varied, and the DEGs between different samples could be the essential factors related to the pathogenesis and evolution of the AGA.
Functional enrichment analysis
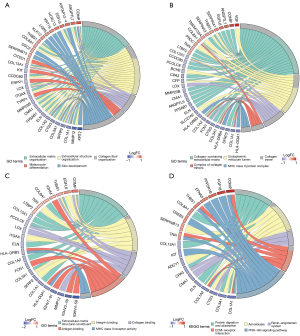
To further delineate the biological functions and related pathways of DEGs, we then performed functional enrichment analysis on the DEGs. The GO annotations of DEGs mainly enriched in the extracellular matrix (ECM)-related pathways (Figure 2A-2C), such as ECM organization, collagen-containing ECM, and ECM structural constituent. Intriguingly, collagen-related terms were also implicated, including collagen fibril organization, collagen trimer, and collagen binding. Besides, immune-related activities were also involved in the functional enrichment of DEGs, spanning MHC class II protein complex, antigen binding, and MHC class II receptor activity. Next, KEGG analysis was used to investigate the associated enrichment pathway of DEGs (Figure 2D), with results demonstrating that DEGs were significantly enriched in protein digestion and absorption, amoebiasis, renin-angiotensin system, ECM-receptor interaction, and the PI3K-Akt signaling pathway. Summarily, DEGs between AGA and healthy individuals disclosed that the complex mechanisms of AGA, simultaneously elucidating that the DEGs are the nexus of immune cells and immune-related process or activities.
Screening of key genes of AGA
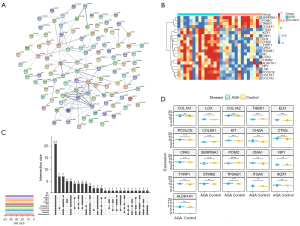
To screen the key genes in the DEGs, the PPI network was constructed using the STRING database with minimum required interaction score of 0.4 (Figure 3A), thus obtaining an interaction network file. Next, 10 algorithms in the cytoHubba of Cytoscape, as previously described, were used to calculate and rank the score of each gene. We then intersected the top 50 genes obtained in the 10 algorithms using UpSet plot (Figure 3B), thereby identifying shared 21 genes, which included SERPINA1, POMC, THBS1, COL8A1, ITGAX, IKZF1, CMA1, ELN, CHGA, TYRP1, CPA3, TPSAB1, KIT, CTSG, STMN2, ALDH1A1, NPY, LOX, PCOLCE, COL1A1, and COL1A2. The expression levels of key genes of the AGA patients and healthy controls were visualized using a heatmap (Figure 3C), and presented specific transcriptomic patterns in AGA patients, manifesting augmented levels in most of the aforementioned key genes. Specifically, SERPINA1, POMC, THBS1, and COL8A1 were significantly highly expressed in the samples originated from scalp skin vertex biopsies of healthy volunteers, and enhanced expression of remaining 17 genes was presented in the AGA patient samples (Figure 3D). Summarily, we could tentatively propose that such 21 key genes may assume key functions in the developments of the AGA, and, more importantly, could be used as a potential diagnostic biomarker as well as promising targets tailoring therapeutic regimens in the managements of AGA.
Immune cell infiltration analysis of AGA and healthy scalp skin
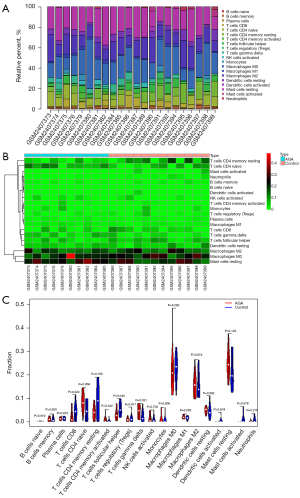
Next, we aimed to parse the immune cell infiltration in AGA and healthy scalp skin using the CIBERSORT algorithm. After excluding the samples with P>0.05, 10 AGA and 12 healthy samples were retained for the following analyses. Relative abundances of 20 kinds of immune cells in these samples were presented in histogram (Figure 4A), with the heatmap showing the normalized absolute abundance (Figure 4B) of immune cells. Notably, memory resting CD4 T cells, naive CD4 T cells, M2 macrophages, M0 macrophages, and resting mast cells were the leading types of infiltrating immune cells in the sample. We further compared the relative abundances of immune cells between AGA patients and healthy controls using Wilcoxon test, among which the follicular helper T cells manifested a higher relative abundance in control group, and a significant increase of gamma delta T cells in AGA patients compared with healthy individuals was presented (Figure 4C). Here, we discovered that the relative abundances of certain types of immune cells varies, coupled with the immune-related terms obtained by the functional enrichment analysis of key genes, indicating that the key genes play an essential role in the pathogenesis of AGA, whereby possibly influencing the relative abundance of immune cells and immune-related activity.
Identifications of IRGs
As previously described, we maintained that the immune cells and immune-related activities could contribute to the developments of AGA, and the aforementioned key genes may act as a connecting bridge to the immune system and phenotype changes of AGA in cellular levels.
To confirm this hypothesis, we further delineated the correlation between the key genes and varying immune cell subtypes. Notably, the correlation analysis disclosed the close associations among key genes, and their relationships with immune cells (Figure 5A). Specifically, we found that certain key genes are significantly correlated with multiple immune cells, except for ALDH1A1, COL8A1, ELN, POMC, STMN2, and THBS1 (Figure 5B), which were termed as IRGs. Among which, mast cells presented the most significant associations with the IRGs, with activated and resting mast cells significantly correlated with seven and eight IRGs, respectively, followed by follicular helper T cells. Thus, we postulated that the 15 IRGs are the essential indicators reflecting the immune component remodeling in the local microenvironment, and the potential diagnostic biomarker and therapeutic targets in AGA. The following enrichment analyses revealed that the 15 IRGs are mainly enriched in the collagen-related GO terms (Figure 5C), such as fibrillar collagen trimer in cellular component and collagen-binding in molecular function. Besides, hormone-related terms (e.g., peptide hormone processing) and endocrine-related terms (e.g., platelet-derived growth factor binding) are also involved. Interestingly, the KEGG analysis suggested that the immune-related pathway, complement and coagulation cascades, was associated (Figure 5D). Summarily, we noticed that the 15 genes that significantly related to the relative abundances of various immune cell subtypes were predominantly enriched in the immune- and collagen-related GO terms, and further indicated that such genes could be candidate genes involved in the pathogenesis of AGA.
Identification and validation of hub genes for AGA
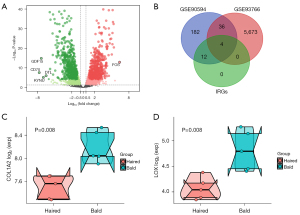
It is well documented that DPCs are pivotal in the growth process of hair, and cultured DPCs have been used as a cellular model to investigate the causative factors for AGA (23-25). Thus, the high throughput sequencing data of GSE93766, containing immortalized balding and non-balding human DPCs derived from male AGA patients, was used for the following analysis, thus identifying the hub genes for AGA combing with GSE90594. First, the identification of DEGs between immortalized balding and non-balding DPCs revealed a total of 5,797 DEGs, and P<0.05 and |logFC)| >0.5 were set as the threshold (Figure 6A). Notably, FOS presented the greatest logFC value in up-regulation DEGs, while CD70 was most down-regulated gene in AGA samples. Next, we intersected the DEGs obtained from GSE93766 and GSE90594 with 15 IRGs, thus identifying four hub genes for AGA, comprising COL1A2, PCOLCE, ITGAX, and LOX (Figure 6B). Next, to further validate the results, GSE36169 was used to confirm the hub genes, which contained expression profiling of biopsied from haired and bald scalp of five individuals with AGA. Specifically, the expression levels of COL1A2 and LOX were significantly diminished in the haired scalp of AGA patients (Figure 6C,6D), which agreed with the previous result. Notwithstanding, PCOLCE and ITGAX were not detected in this dataset, but this may have been due to discrepancies in sequencing depth and sample biases.
Discussion
AGA is a multifactorial disease, featured by typical clinical manifestations and high prevalence in both genders (26). To date, the essential factors implicated in the etiopathogenesis of AGA have been well characterized, represented by dihydrotestosterone (DHT) (27,28), genetic predisposition (29,30), environmental input (31), and aging (3,25). Minoxidil and finasteride remain the only two kinds of pharmacological therapies approved by the FDA (8), with encouraging efficacy presenting in the clinical settings (32). Nevertheless, their treatment responses have varied (31,33), accompanied by various degrees of adverse reactions (34). Despite tremendous progress having been achieved concerning the pathogenesis and managements of AGA in the past decades (31), it is unequivocal from the available literature that much remains to be addressed, especially in the identification of the alternative or individualized therapeutic approaches with lessened side effects and improved efficacy (1,5).
The HF is a delicate organ undergoing cyclic involution and regeneration (35,36), and HFSCs are indispensable part of the hair regenerations (37), supported by the varying nearby cells in the local environment, together constituting a HFSC niche (17). Notably, except for HFSC itself, the niche consists of various cell types, including DPCs, adipocytes, and immune cells (17). Among them, DPCs take center stage in the activations of HFSCs, thereby providing a pivotal signal initiating hair regeneration and orchestrating the crosstalk among disparate cell types in the microenvironment (38). Intriguingly, androgen receptors (AR) are predominantly expressed by DPCs, but not expressed by keratinocytes in the HF (17), with compelling evidence indicating that significantly elevated AR expressions can be observed in the DPCs originated from bald scalp compared to the non-bald scalp samples of AGA patients (27), which was also supported by in vitro data (39). Cellular actors of DPCs implicated in physiologic and pathophysiologic circumstances make it an ideal model investigating the underlying mechanism of AGA (25,38).
In our study, high throughput sequencing expression profiling of immortalized balding and non-balding DPCs derived from male AGA patients, combining with the RNA-seq data of clinical biopsies of AGA and healthy volunteers, was used to identify the hub genes for AGA, comprising COL1A2, PCOLCE, ITGAX, and LOX. Note worthily, the role of COL1A2 in HF has been appreciated, which belongs to collagens family members (40,41), participating in the formations of ECM, with conclusive reports maintaining that the cross-talk between ECM and dermal papilla is indispensable for the HF development and regeneration (42), and that LOX also plays a similar yet specific role that is responsible for deposition and stabilization of ECM, which is regulated by transforming growth factor (TGF)-β signaling pathways, a pivotal player in the control of HF developmental and AGA pathogenesis (31,43). Similarly, PCOLCE is an essential factor in the telogen-to-anagen transition of HF (44), manifesting enhanced expression levels in DPCs and also known as an activator of BMP1 in bone morphogenetic protein (BMP) signaling pathways (41). Nevertheless, the associations with ITGAX and AGA remain elusive, with many lines of evidence reveling that ITGAX is a candidate gene for autoimmune diseases (45), which may shed the light on the fact that autoimmune mechanisms may be implicated in AGA, like alopecia areata (46). Collectively, we could discover that the hub genes identified in this study are closely linked with the causative factors of AGA, which could be used as the viable diagnostic and therapeutic target in the clinical applications.
In recent times, attention has increasingly been given to the functions of immune cells in the skin. In addition to assuming nascent functions of immune surveillance and microbial defense, immune cells are integral parts of homeostasis maintenance of skin (18), hair cycling induction (19), and facilitating wound healing (47). More importantly, immune cells have been demonstrated to contribute to immune-mediated alopecias in humans, such as alopecia areata (48), primary cicatricial alopecia (49), and frontal fibrosing alopecia (50). Immune profiles of such types of alopecia have been well delineated before, thus offering novel avenues for exploration in future research (49). Notwithstanding, as research into the role of the immune cell regulation in AGA is in its infancy, further studies into this field are still imperative, thereby breaking through the present bottle necks.
In this study, we aimed to identify the potential therapeutic targets of AGA using multiple datasets in the GEO database, and we discovered that the DEGs between the scalp skin vertexes of male AGA patients and healthy volunteers were specifically enriched in the immune-related terms. The following analysis based on the CIBERSORT algorithm indicated that the AGA patients presented disparate relative immune cell composition to that of the healthy volunteers, and high abundance of CD4 T cells, macrophages, and mast cells were consistently presented in both diseased and healthy scalps. Thus, we assert that immune-related activities and immune inflammatory cells contribute to the development and progression of AGA, as previously reported (51,52). However, whether the immune cells are associated with the typical phenotypes of AGA remain uncharacterized. In this regard, further study is warranted. This study has certain limitations. First, the sample sizes of datasets used in the study were comparatively small, and larger sample-sized, or multicentered cohorts are needed to increase the reliability of these conclusions. Second, it is imperative to validate our findings through ongoing experiments.
Conclusions
In this study, we used high throughput sequencing expression profiling of immortalized balding and non-balding DPCs, combining with the RNA-seq data of clinical biopsies of AGA and healthy volunteers, thus identifying the hub genes for AGA. Besides, we also dissected the immune cell infiltration of AGA, providing novel perspectives for the immune mechanism of AGA.
Acknowledgments
Funding: This study was funded by Young Scholars of The People’s Hospital of Guangxi Zhuang Autonomous Region (No. QN2020-23).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-4634/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4634/coif). All authors report that this study was funded by Young Scholars of The People’s Hospital of Guangxi Zhuang Autonomous Region (No. QN2020-23). The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lolli F, Pallotti F, Rossi A, et al. Androgenetic alopecia: a review. Endocrine 2017;57:9-17. [Crossref] [PubMed]
- Redler S, Messenger AG, Betz RC. Genetics and other factors in the aetiology of female pattern hair loss. Exp Dermatol 2017;26:510-7. [Crossref] [PubMed]
- Xiong J, Wu B, Hou Q, et al. Comprehensive Analysis of LncRNA AC010789.1 Delays Androgenic Alopecia Progression by Targeting MicroRNA-21 and the Wnt/β-Catenin Signaling Pathway in Hair Follicle Stem Cells. Front Genet 2022;13:782750. [Crossref] [PubMed]
- Yip L, Zaloumis S, Irwin D, et al. Gene-wide association study between the aromatase gene (CYP19A1) and female pattern hair loss. Br J Dermatol 2009;161:289-94. [Crossref] [PubMed]
- Motofei IG, Rowland DL, Baconi DL, et al. Androgenetic alopecia; drug safety and therapeutic strategies. Expert Opin Drug Saf 2018;17:407-12. [Crossref] [PubMed]
- Su LH, Chen LS, Chen HH. Factors associated with female pattern hair loss and its prevalence in Taiwanese women: a community-based survey. J Am Acad Dermatol 2013;69:e69-77. [Crossref] [PubMed]
- Varothai S, Bergfeld WF. Androgenetic alopecia: an evidence-based treatment update. Am J Clin Dermatol 2014;15:217-30. [Crossref] [PubMed]
- Adil A, Godwin M. The effectiveness of treatments for androgenetic alopecia: A systematic review and meta-analysis. J Am Acad Dermatol 2017;77:136-41.e5. [Crossref] [PubMed]
- Stoffel-Wagner B. Neurosteroid biosynthesis in the human brain and its clinical implications. Ann N Y Acad Sci 2003;1007:64-78. [Crossref] [PubMed]
- Irwig MS, Kolukula S. Persistent sexual side effects of finasteride for male pattern hair loss. J Sex Med 2011;8:1747-53. [Crossref] [PubMed]
- Rebora A. Telogen effluvium: a comprehensive review. Clin Cosmet Investig Dermatol 2019;12:583-90. [Crossref] [PubMed]
- Kamyab K, Rezvani M, Seirafi H, et al. Distinguishing immunohistochemical features of alopecia areata from androgenic alopecia. J Cosmet Dermatol 2019;18:422-6. [Crossref] [PubMed]
- Matsumura H, Mohri Y, Binh NT, et al. Hair follicle aging is driven by transepidermal elimination of stem cells via COL17A1 proteolysis. Science 2016;351:aad4395. [Crossref] [PubMed]
- Vasserot AP, Geyfman M, Poloso NJ. Androgenetic alopecia: combing the hair follicle signaling pathways for new therapeutic targets and more effective treatment options. Expert Opin Ther Targets 2019;23:755-71. [Crossref] [PubMed]
- Castellana D, Paus R, Perez-Moreno M. Macrophages contribute to the cyclic activation of adult hair follicle stem cells. PLoS Biol 2014;12:e1002002. [Crossref] [PubMed]
- Ali N, Zirak B, Rodriguez RS, et al. Regulatory T Cells in Skin Facilitate Epithelial Stem Cell Differentiation. Cell 2017;169:1119-29.e11. [Crossref] [PubMed]
- Chen CL, Huang WY, Wang EHC, et al. Functional complexity of hair follicle stem cell niche and therapeutic targeting of niche dysfunction for hair regeneration. J Biomed Sci 2020;27:43. [Crossref] [PubMed]
- Muneeb F, Hardman JA, Paus R. Hair growth control by innate immunocytes: Perifollicular macrophages revisited. Exp Dermatol 2019;28:425-31. [Crossref] [PubMed]
- Wang ECE, Higgins CA. Immune cell regulation of the hair cycle. Exp Dermatol 2020;29:322-33. [Crossref] [PubMed]
- Garza LA, Yang CC, Zhao T, et al. Bald scalp in men with androgenetic alopecia retains hair follicle stem cells but lacks CD200-rich and CD34-positive hair follicle progenitor cells. J Clin Invest 2011;121:613-22. [Crossref] [PubMed]
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453-7. [Crossref] [PubMed]
- Chen S, Sun Y, Zhu X, et al. Prediction of Survival Outcome in Lower-Grade Glioma Using a Prognostic Signature with 33 Immune-Related Gene Pairs. Int J Gen Med 2021;14:8149-60. [Crossref] [PubMed]
- Inui S, Itami S. Androgen receptor transactivity is potentiated by TGF-β1 through Smad3 but checked by its coactivator Hic-5/ARA55 in balding dermal papilla cells. J Dermatol Sci 2011;64:149-51. [Crossref] [PubMed]
- Shin H, Yoo HG, Inui S, et al. Induction of transforming growth factor-beta 1 by androgen is mediated by reactive oxygen species in hair follicle dermal papilla cells. BMB Rep 2013;46:460-4. [Crossref] [PubMed]
- Upton JH, Hannen RF, Bahta AW, et al. Oxidative stress-associated senescence in dermal papilla cells of men with androgenetic alopecia. J Invest Dermatol 2015;135:1244-52. [Crossref] [PubMed]
- Kelly Y, Blanco A, Tosti A. Androgenetic Alopecia: An Update of Treatment Options. Drugs 2016;76:1349-64. [Crossref] [PubMed]
- Batrinos ML. The endocrinology of baldness. Hormones (Athens) 2014;13:197-212. [Crossref] [PubMed]
- Rathnayake D, Sinclair R. Male androgenetic alopecia. Expert Opin Pharmacother 2010;11:1295-304. [Crossref] [PubMed]
- Heilmann-Heimbach S, Herold C, Hochfeld LM, et al. Meta-analysis identifies novel risk loci and yields systematic insights into the biology of male-pattern baldness. Nat Commun 2017;8:14694. [Crossref] [PubMed]
- Heilmann-Heimbach S, Hochfeld LM, Paus R, et al. Hunting the genes in male-pattern alopecia: how important are they, how close are we and what will they tell us? Exp Dermatol 2016;25:251-7. [Crossref] [PubMed]
- Guo H, Gao WV, Endo H, et al. Experimental and early investigational drugs for androgenetic alopecia. Expert Opin Investig Drugs 2017;26:917-32. [Crossref] [PubMed]
- York K, Meah N, Bhoyrul B, et al. A review of the treatment of male pattern hair loss. Expert Opin Pharmacother 2020;21:603-12. [Crossref] [PubMed]
- Rossi A, Cantisani C, Scarnò M, et al. Finasteride, 1 mg daily administration on male androgenetic alopecia in different age groups: 10-year follow-up. Dermatol Ther 2011;24:455-61. [Crossref] [PubMed]
- Jain R, De-Eknamkul W. Potential targets in the discovery of new hair growth promoters for androgenic alopecia. Expert Opin Ther Targets 2014;18:787-806. [Crossref] [PubMed]
- Chen CC, Plikus MV, Tang PC, et al. The Modulatable Stem Cell Niche: Tissue Interactions during Hair and Feather Follicle Regeneration. J Mol Biol 2016;428:1423-40. [Crossref] [PubMed]
- Hsu YC, Li L, Fuchs E. Emerging interactions between skin stem cells and their niches. Nat Med 2014;20:847-56. [Crossref] [PubMed]
- Hsu YC, Pasolli HA, Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell 2011;144:92-105. [Crossref] [PubMed]
- Ceruti JM, Oppenheimer FM, Leirós GJ, et al. Androgens downregulate BMP2 impairing the inductive role of dermal papilla cells on hair follicle stem cells differentiation. Mol Cell Endocrinol 2021;520:111096. [Crossref] [PubMed]
- Inui S, Fukuzato Y, Nakajima T, et al. Androgen-inducible TGF-beta1 from balding dermal papilla cells inhibits epithelial cell growth: a clue to understand paradoxical effects of androgen on human hair growth. Faseb j 2002;16:1967-9. [Crossref] [PubMed]
- Ge W, Tan SJ, Wang SH, et al. Single-cell Transcriptome Profiling reveals Dermal and Epithelial cell fate decisions during Embryonic Hair Follicle Development. Theranostics 2020;10:7581-98. [Crossref] [PubMed]
- Li C, Li Y, Zhou G, et al. Whole-genome bisulfite sequencing of goat skins identifies signatures associated with hair cycling. BMC Genomics 2018;19:638. [Crossref] [PubMed]
- Wang J, Sui J, Mao C, et al. Identification of Key Pathways and Genes Related to the Development of Hair Follicle Cycle in Cashmere Goats. Genes (Basel) 2021;12:180. [Crossref] [PubMed]
- Inui S, Itami S. Molecular basis of androgenetic alopecia: From androgen to paracrine mediators through dermal papilla. J Dermatol Sci 2011;61:1-6. [Crossref] [PubMed]
- Zhang X, Bao P, Ye N, et al. Identification of the Key Genes Associated with the Yak Hair Follicle Cycle. Genes (Basel) 2021;13:32. [Crossref] [PubMed]
- Shi D, Zhong Z, Xu R, et al. Association of ITGAX and ITGAM gene polymorphisms with susceptibility to IgA nephropathy. J Hum Genet 2019;64:927-35. [Crossref] [PubMed]
- Betz RC, Petukhova L, Ripke S, et al. Genome-wide meta-analysis in alopecia areata resolves HLA associations and reveals two new susceptibility loci. Nat Commun 2015;6:5966. [Crossref] [PubMed]
- Komi DEA, Khomtchouk K, Santa Maria PL. A Review of the Contribution of Mast Cells in Wound Healing: Involved Molecular and Cellular Mechanisms. Clin Rev Allergy Immunol 2020;58:298-312. [Crossref] [PubMed]
- Xing L, Dai Z, Jabbari A, et al. Alopecia areata is driven by cytotoxic T lymphocytes and is reversed by JAK inhibition. Nat Med 2014;20:1043-9. [Crossref] [PubMed]
- Anzai A, Wang EHC, Lee EY, et al. Pathomechanisms of immune-mediated alopecia. Int Immunol 2019;31:439-47. [Crossref] [PubMed]
- Del Duca E, Ruano Ruiz J, Pavel AB, et al. Frontal fibrosing alopecia shows robust T helper 1 and Janus kinase 3 skewing. Br J Dermatol 2020;183:1083-93. [Crossref] [PubMed]
- Magro CM, Rossi A, Poe J, et al. The role of inflammation and immunity in the pathogenesis of androgenetic alopecia. J Drugs Dermatol 2011;10:1404-11. [PubMed]
- Young JW, Conte ET, Leavitt ML, et al. Cutaneous immunopathology of androgenetic alopecia. J Am Osteopath Assoc 1991;91:765-71. [Crossref] [PubMed]
(English Language Editor: J. Jones)


