
Single-cell transcriptome analysis of tumor immune microenvironment characteristics in colorectal cancer liver metastasis
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors worldwide. According to the 2020 report of the International Agency for Research on Cancer (IARC), the morbidity and mortality associated with CRC ranks third and second among all cancers, respectively (1). In China, CRC is the second most common malignancy, with 560,000 new cases and 290,000 deaths reported in 2020, and the disease burden continues to increase dramatically. Early-stage CRC can achieve a better outcome through radical surgery, but the prognosis of metastatic CRC (mCRC) is poor due to the lack of effective treatment, with a 5-year survival rate of less than 10%. Liver is the most common distant metastatic organ of CRC. The probability of synchronous liver metastasis in CRC is about 20–25%, and the proportion of liver metastasis in the whole course of the disease is as high as 40–50%. Of all patients who died from CRC, 49% had liver-predominant lesions, and 83% had liver involvement (2). Therefore, liver metastasis is the leading cause of death in CRC patients.
Malignant cells in tumors interact with immune cells and non-immune cells to form a complex cellular network known as the tumor microenvironment (TME) (3). To date, the mechanisms of liver metastasis in CRC remain unclear. In addition, factors related to cancer cells themselves, alteration in the TME may also play an important role, including tumor-related inflammatory responses and micro-angiogenesis. The ability to establish secondary tumors in liver depends on the interaction of tumor cells with the specific microenvironmental factors in the liver. These microenvironmental factors play a bidirectional role in inhibiting or promoting metastasis and growth (4). CRC metastasis begins when tumor cells shed from the primary site and enter the circulation, with the liver being the first filter for circulating tumor cells (5,6). The majority of tumor cells are arrested by the pro-inflammatory response induced by liver sinusoidal endothelial cells (LSECs), Kupffer cells (KCs), and lymphocytes in the hepatic sinusoid (7), However, a few survivor cells can infiltrate through specialized endothelial barriers into the liver parenchyma. Hepatocytes are an important source of pro-inflammatory signals during metastasis and cooperate with lymphocytes and myeloid cells to establish a pro-metastatic niche (8). The recruitment and activation of stromal cells further contribute to the pro-metastatic niche and promote angiogenesis, providing an adequate blood supply for metastasis. Tumor growth-promoting factors secreted by neighboring hepatocytes, hepatic stem cells, and tumor-associated macrophages (TAMs) drive micro-metastasis to macro-metastasis (9,10).
Hepatic macrophages can be induced to kill tumor cells in the early stages of CRC liver metastasis, and can also be induced to generate tumor-promoting functions necessary for macro-metastasis (11,12), through the macrophage polarization system including the “classically activation” M1 phenotype and the “alternately activation” M2 phenotype (13). M1-TAMs induce inflammatory responses to generate reactive oxygen and nitrogen radicals, which can change cells and the surrounding microenvironment, thereby promoting tumor progression. M2-TAMs suppress tumor immune responses and destroy the immune barrier by secreting immunosuppressive cytokines, resulting in the imbalance of defense mechanisms. In addition, M2-TAMs secrete growth factors, including transforming growth factor (TGF) and vascular endothelial growth factor (VEGF), to promote tumor proliferation and extracellular matrix (ECM) remodeling. Furthermore, M2-TAMs can induce the recruitment and activation of fibroblasts, pericytes and endothelial cells to drive angiogenesis within metastasis (9). This complex and diverse mechanism not only promotes tumor colonization in “soil”, but also changes the local “soil” microenvironment to make it suitable for “seed” growth.
Over the past decade, the TME has been a hot area of tumor biology research and is considered a new target for cancer therapy. Immune checkpoint blockade therapy has achieved major breakthroughs in many malignancies, and programmed cell death protein 1 (PD-1) inhibitors have become the standard therapy in microsatellite instability-high (MSI-H) CRC. However, proficient mismatch repair (pMMR)/non-MSI-H CRC does not benefit from immune checkpoint inhibitor (ICIs) alone (14), rather, ICIs combined with anti-angiogenic agents have shown potential to reverse a subset of pMMR/non-MSI-H mCRC that is unresponsive to ICIs (15). Tumor heterogeneity may lead to different responses in patients undergoing the same treatment. CRC is one of the less immune-infiltrating cancers, which may account for the poor response to anti-PD-1 therapy. Generally, the heterogeneity of tumor cells is more emphasized than other components in the TME (16). In fact, there is also heterogeneity in stromal cells, including tumor-infiltrating immune cells (17). Liver metastasis of CRC is a complex biological process involving multiple factors and steps, and its mechanisms remain to be fully elucidated. Tumor-stroma interactions in the TME can promote cancer invasion and metastasis through chemokine signaling. The TME contains many host cells that may suppress or promote the aggressiveness of cancers. Several host-derived myeloid cells are present in the TME and their recruitment is controlled by chemokine signaling (18). Recently, in addition to immunotherapeutic strategies targeting PD-1/PD-1 ligand (PD-L1) or cytotoxic T-lymphocyte-associated protein 4 (CTLA-4), TAMs and cancer-associated fibroblasts (CAFs) have also been found to be therapeutic targets (19,20). Blocking the recruitment of TAMs or depleting the population of TAMs, and inducing macrophage polarization from the immunosuppressive M2 to the pro-inflammatory and cytotoxic M1 may be viable therapeutic strategies.
Tumors are complex ecosystems. In the past, due to the lack of accurate and complete understanding of the TME, it was impossible to translate precise and effective treatment strategies into clinical practice (21-23). Due to the low-resolution landscape, traditional bulk RNA sequencing could not specifically represent the changes in cell populations in the TME (24,25). Single-cell RNA sequencing (scRNA-seq) is a technique for transcriptome analysis at the single-cell level, enabling a comprehensive and detailed characterization of cellular diversity and heterogeneous phenotypes (17,26). In addition, since most studies compared the phenotypic differences between CRC primary tumor and liver metastasis in different patients, differences in individual genetic characteristics meant that it was not possible to accurately reflect the transition of the TME from primary tumor to metastasis. In this study, scRNA-seq was performed on the primary tumor and the metastatic liver tumor from the same patient to identify the altered characteristics of the TME in synchronous liver metastasis of CRC, and to reveal its molecular regulatory mechanisms. This data will provide a theoretical basis for immunotherapy in patients with mCRC. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5270/rc).
Methods
Sample collection
Primary colon cancer and liver metastatic samples were obtained from a 71-year-old male patient. He was diagnosed with sigmoid colon cancer with simultaneous liver metastasis in March 2021, and underwent laparoscopic radical sigmoidectomy combined with liver tumor resection on March 12, 2021. The malignancy was confirmed by pathology. The samples used in this study were from tumors removed during surgery. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of The Third Affiliated Hospital of Soochow University (No. 2022-science-158). The ethics committee approved this study to be exempt from the informed consent process.
ScRNA-seq
Cell capture was performed using the Single Cell 5' Library and Gel Bead Kit and the Chromium Single Cell A Chip Kit (10x Genomics, CA USA), according to the manufacturer's protocol. Captured cells were lysed and the released RNA was barcoded by reverse transcription in individual Gel Bead in Emulsions (GEMs). The complementary DNA (cDNA) was generated, amplified, and quality assessed. Subsequently, the scRNA-seq library was constructed, as previously described (27,28). Finally, the library was sequenced using an Illumina Novaseq6000 sequencer. The sequencing depth was at least 100,000 reads per cell, using a pair-end 150 bp (PE150) reading strategy.
ScRNA-seq data processing
ScRNA-seq fastq.gz files obtained from 10x Genomics were aligned and quantified using the hg38 human reference genome and CellRanger software. The filtered data were import into the R software by Read10x function packaged in R package Seurat (Version 3.2.3). The DoubletFinder package pipeline was used to remove doublets. For further quality control, cells with less than 8000 unique molecular identifier (UMI) count, less than 75% mitochondrial gene count, and less than 45% ribosomal gene count were used for further analysis.
ScRNA-seq integration, dimension reduction, and unsupervised clustering
The workflow in Seurat was followed to process single-cell data for dimension reduction and unsupervised clustering. Briefly, the FindVariableFeatures and NormalizeData functions were applied for selecting high-variable genes and Log2 (TPM+1) normalization. Subsequently, to remove batch effect between samples, the IntegrateData function was used to integrate data. The integrated data matrix was used to perform principal component analysis (PCA) and uniform manifold approximation and projection (UMAP) dimension reduction. The top 40 principal components (PCs) were used for downstream analysis using the Elbowplot function of Seurat. Major clusters were identified by FindClusters function (resolution =0.5). Each cluster was categorized into a known biological cell type according to previously identified cell markers.
Single-cell TCR-seq data processing
ScRNA-seq fastq.gz TCR files obtained from 10x Genomics were aligned and quantified using the hg38 human vdj reference genome and CellRanger software. The filtered csv files generated from CellRanger were matched with paired scRNAseq data for further analysis.
Differential gene expression analysis
Differentially expressed genes between myeloid cells were selected using the edgeR package (version 3.28.1). The CalcNormFactors function was used to normalize raw count data according to the trimmed mean of M-values (TMM) algorithm, and the estimateDisp function was used to estimate the dispersion of gene expression values. Differentially expressed genes were selected and visualized using the ggplot2 and ggrepel packages.
Gene set enrichment analysis
Gene set enrichment analysis was performed using Gene Set Enrichment Analysis (GSEA) software (version 4.1.0), and the gene sets were derived from MSigDB gene sets. The differences in pathway activities scored per cell between clusters were calculated with wilcox.test.
Gene Ontology enrichment analysis
The differentially expressed genes identified using the edgeR package were selected for Gene Ontology (GO) analysis by clusterProfiler R package (29) (Version 3.14.3). The Barplot function was used for visualization.
SCENIC analysis
SCENIC (version 1.2.4) was used to analyze activation regulons in each subset, taking raw count matrix as input. In brief, Gene Network Inference with Ensemble of trees 3 (GENIE3) calculated the co-expression networks and RcisTarget (version 1.6.0) identified the regulons. Subsequently, each cell was scored for regulon activity by AUCell (version 1.13.3). The differentially activated regulons which target TAM2 associated signature genes in each subset were identified using the Wilcox test.
Trajectory analysis of myeloid cells
The Monocle R package (version 2.14.0) that introduced pseudo-time was used to construct single-cell trajectories. Genes that were expressed in more than 10 cells were filtered out.
Calculation of copy number variations
The infercnv R package (version 1.2.1) was used for copy number variation (CNV) assessment based on scRNA-seq raw counts. The hidden Markov model was chosen to predict CNV states. Gene location data were achieved using AnnoProbe (version 1.13.3). CNV-based hierarchical clustering was used to classify subclones of specific subtypes.
Results
Clinical information
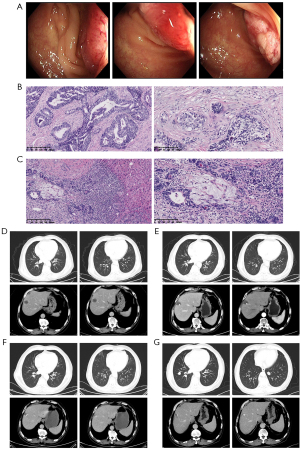
The patient was a 71-year-old male, admitted with lower abdominal pain for 1 month. In March 2021, the following tumor markers were detected: carcinoembryonic antigen (CEA) 75.52 ng/mL and cancer antigen (CA) 19-9 63.56 U/mL. Computed tomography (CT) scan suggested the possibility of metastasis in S8 of the liver, and the patient was subsequently diagnosed with sigmoid colon cancer by colonoscopy. On March 12, 2021, the patient underwent laparoscopic radical sigmoidectomy and liver tumor resection under general anesthesia. Postoperative pathology showed sigmoid colon ulcerative adenocarcinoma that was moderately-poorly differentiated, 3 cm × 2.5 cm in size, infiltration into the subserosa, vessels, and nerves (+), resection margin (−), mesenteric lymph node metastasis (0/10), and liver S8 adenocarcinoma metastasis with necrosis. Gene sequencing revealed a total of 7 somatic variants, 5 of which had clear or potential clinical significance, including: 1. KRAS p.G13D (c.38G>A, p.Gly13Asp 43.04%); 2. APC c.730-1G>A; 3. APC p.M1383fs; 4. SMAD4 p.Y301*; and 5. TP53 p.R282W. The tumor mutational burden (TMB) was 4.99 mutations/Mb and the microsatellite instability detection was microsatellite stable (MSS). No pathogenic germline variants were detected. Postoperative CT scan on April 6, 2021 showed no metastasis. The patient received 8 cycles of Capecitabine plus Oxaliplatin (CAPEOX) adjuvant chemotherapy from April 8 to September 29, 2021, with regular follow-up. CT scan on April 14, 2022 showed that the range of low-density shadows in the liver operation area was larger than before (September 2021), which may be active lesions, and multiple lung metastases occurred. The patient was diagnosed with sigmoid colon cancer postoperatively, with multiple liver and lung metastases. The disease-free survival (DFS) was 13 months. The patient has been receiving first-line palliative treatment with Bevacizumab combined with Irinotecan + Capecitabine since April 21, 2022, and is still under treatment. The colonoscopy, pathology and imaging examinations are shown in Figure 1.
Significant differences in the TME between the primary tumor and the liver metastasis of CRC
Whole tumor 5' single-cell transcriptome and paired single-cell T-cell receptor (TCR) sequencing were performed on the collected primary and metastatic samples of the CRC. After quality control, batch correction, and dimension reduction and unsupervised clustering analysis, 15,438 cells were finally obtained. The cells were divided into 11 populations, including 4 epithelial cell subsets, 2 stromal cell subsets (fibroblasts and endothelial cells), and 5 immune cell subsets [myeloid cells, mast cells, T and natural killer (NK) cells, B cells, and plasma cells] (Figure 2A,2B). Each population was defined by known classical signature genes, such as epithelial cells (KRT8+), fibroblasts (α-SMA+), endothelial cells (CD31+), and immune cells (PTPRC+) (Figure 2B). Subsequently, comparing the proportions of each population in the primary and metastatic samples, it was found that in the immune cell population, the proportion of myeloid cells in the metastatic sample was significantly increased, while the proportion of B cells and plasma cells was significantly decreased. The proportion of the stromal cell population was also significantly reduced in the metastatic tumor. Only two smaller subsets of the epithelial cell population were significantly increased in metastasis (Figure 2C). Cells between different clusters are shown in Table 1.
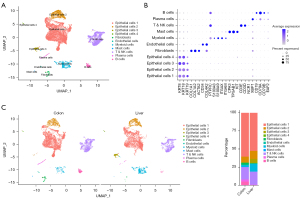
Table 1
| Cells | Colon (num) | Liver (num) |
|---|---|---|
| Epithelial cells 1 | 5,126 | 3,659 |
| Epithelial cells 2 | 689 | 767 |
| Epithelial cells 3 | 8 | 386 |
| Epithelial cells 4 | 23 | 48 |
| Endothelial cells | 52 | 13 |
| Myeloid cells | 208 | 769 |
| Mast cells | 28 | 5 |
| T & NK cells | 1,450 | 1,253 |
| Plasma cells | 249 | 29 |
| B cells | 423 | 22 |
NK, natural killer.
Differences in epithelial cell function between primary and metastatic lesions
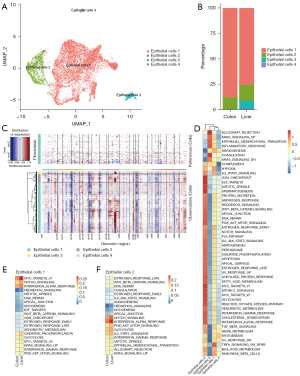
Epithelial cells were roughly divided into four subgroups (named epithelial cells 1–4). Epithelial cells 2 showed high expression of proliferation-related genes such as MKI67, indicating that they were in a highly proliferative state. Epithelial cells 3 were mainly found in metastasis (Figure 3A,3B). CNV analysis was performed on all epithelial cells, and the results showed that epithelial cells 3 had a higher degree of CNV, which may represent a higher degree of malignancy, while epithelial cells 1 and 2 had a relatively low degree of copy number variation, which may suggest a transitional stage of malignant transformation (Figure 3C). Subsequently, functional enrichment analysis was performed on the four subgroups to compare functional differences between the different epithelial cells. The results showed that pathways related to cell cycle and metabolism were significantly enriched in epithelial cells 2, while KRAS and epithelial-mesenchymal transition (EMT) pathways were significantly enriched in epithelial cells 3 (Figure 3D). Subsequently, the functional differences of epithelial cells 1 and 2 between primary and metastatic lesions were further analyzed, and it was found that the NOTCH signaling pathway of epithelial cells 1 and 2 was significantly upregulated in metastasis (Figure 3E,3F).
Myeloid-derived suppressor cells (MDSCs) are significantly enriched in liver metastasis of CRC
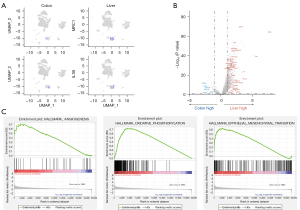
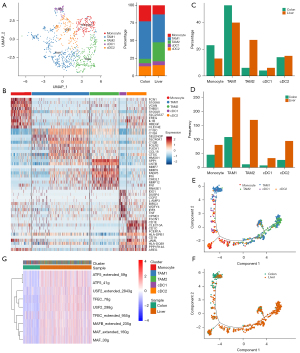
Single-cell sequencing results showed that the proportion of myeloid cells varied most significantly in the immune cell population. Therefore, we further analyzed the differences in myeloid cells between primary and metastatic samples. By comparing the expression of the MDSC characteristic gene mannose receptor C-type 1 (MRC1)/CD206 between the primary and metastatic lesions, it was found that MRC1 was significantly upregulated in metastatic lesions, while IL1B was highly expressed in primary lesions (Figure 4A). Differentially expressed gene analysis showed that TAM2-related genes (SPP1, CCL18, CXCL9, CCL5, and CCL2) and some genes related to lipid metabolism and extracellular matrix were highly expressed in metastasis (Figure 4B). Gene function enrichment analysis revealed that the functions of angiogenesis, oxidative phosphorylation, and EMT of myeloid cells in the liver metastasis were significantly enhanced (Figure 4C).
Myeloid cells were further divided into 5 subgroups: monocytes, TAM1, TAM2, cDC1, and cDC2 (Figure 5A,5B). Analysis of the proportion of each subgroup showed that the number of myeloid cells in the primary sample was relatively small, and mainly monocytes and TAM1; while the proportion and number of TAM2 were significantly upregulated in the metastatic sample (Figure 5C,5D). Pseudotime analysis also found that myeloid cells showed different differentiation directions in the primary and metastatic lesions, with tumor-promoting myeloid cells TAM2 mainly observed in the metastatic sample (Figure 5E,5F). The transcriptional regulatory network was constructed using SCENIC (30) and the differences in the level of transcriptional regulation between primary and metastatic samples were analyzed. The results showed that transcription factors activating transcription factor 5 (ATF5), transcription factor EC (TFEC), upstream transcription factor 2 (USF2), macrophage activating factor (MAF), and MAFB, which regulate the signature genes of TAM2, had significantly increased activities in metastasis (Figure 5E-5G).
T cells are exhausted in colon cancer liver metastasis
The T cells and NK cells were then divided into five subsets, namely, CD4+, CD8+, regulatory T cells (Tregs), NK, and proliferating T cells (Figure 6). Analysis of the proportions of different subsets revealed that the proportion of CD4+ T cells in the metastatic cancer increased significantly, while the proportion of CD8+ T cells decreased. Other subgroups accounted for a smaller proportion and no significant changes were detected (Figure 6B). Single-cell transcriptome sequencing-matched TCR-sequencing data showed a higher proportion of clone expansion of CD4+ T cells in the metastatic sample, while that of CD8+ T cells was lower (Figure 6D,6E). However, when calculating the average number of T cells that underwent clone expansion, it was found that a subset of CD8+ T cells and Tregs had higher levels of clone expansion in the metastatic tumor (Figure 6F), but no obvious antitumor immune response was found in this subset of CD8+ T cells. The expression of immune checkpoint molecules and effectors of T cell subsets in primary and metastatic samples was further compared. The results showed that PD-1, T cell immunoglobulin domain and mucin domain-3 (TIM-3), and Lymphocyte Activation Gene-3 (LAG-3) expression increased in metastatic lesions compared to primary lesion (Figure 6G).
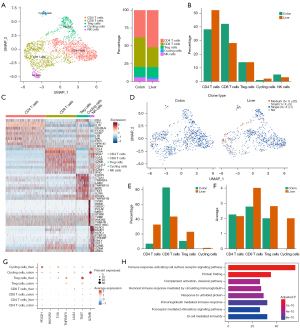
GO enrichment analysis on genes that were downregulated by CD4+ T cells in metastasis revealed that the pathways related to humoral immunity (HI) and B cell-mediated immunity were significantly enriched (Figure 6H).
Intercellular communication analysis
The CellphoneDB package (31) was used to analyze the interaction network between the cell populations identified. Correlation heatmap between cell populations showed stronger interactions between myeloid cells and other cell populations (Figure 7A). Myeloid cells typically secrete high levels of chemokines with the ability to induce directed chemotaxis of nearby responding cells. Therefore, we further analyzed the chemokine interactions between myeloid cells and other cell populations. The results showed that endothelial cells and fibroblasts expressed CXCL12 which acted on CXCR4 of myeloid cells, and T and NK cells interacted with myeloid cells through CCL5-CCR1/CCR5, CCL4-CCR5, and CCL4L2-VSIR. In contrast, myeloid cells expressed CCL4L2, which acted on the progesterone receptor (PGRMC2) on endothelial cells, plasma cells, and fibroblasts, and interacted with T and NK cells via CXCL16-CXCR6 and CXCL9-CXCR3. Myeloid cells expressed CCL4, CCL3, and CCL4L2, which acted on solute carrier family 7 member 1 (SLC7A1), insulin degrading enzyme (IDE), and V-set immunoregulatory receptor (VSIR) on epithelial cells, respectively. Furthermore, interaction with endothelial cells was mediated by CXCL8/CCL5-atypical chemokine receptor 1 (ACKR1), CCL3-IDE, and CCL4L2-VSIR; interaction with fibroblasts, T & NK cells, and plasma cells was mediated through CCL4L2-VSIR; and interaction with B cells was mediated through CCL4L2-VSIR and CCL4-cannabinoid receptor 2 (CNR2) (Figure 7B).

In terms of cell proliferation and angiogenesis, epithelial cells acted on myeloid cells through heparin-binding epidermal growth factor (HBEGF)-CD44, VEGFA- neuropilin 1 (NRP1)/NRP2, and VEGFB-NRP1. Endothelial cells expressed growth factors such as transforming growth factor beta 1 (TGFB1), platelet-derived growth factor beta (PDGFB), insulin-like growth factors 2 (IGF2), placental growth factor (PGF), and HBEGF to interact with TGFB receptor 1 (TGFBR1), LRP1, IGF2 receptor (IGF2R), fms-like tyrosine kinase 1 (FLT1)/NRP2/NRP1, and CD44 on myeloid cells, respectively. Fibroblasts acted on myeloid cells through IGF2-IGF2R, hepatocyte growth factor (HGF)-CD44, PGF-FLT1/NRP2/NRP1, and VEGFA-NRP2/NRP1. Conversely, myeloid cells expressed HBEGF, which acted on CD44 on almost all cells except endothelial cells, such as epithelial cells, fibroblasts, mast cells, T & NK cells, B cells, and plasma cells, and interacted with fibroblasts through PDGFC-PDGF receptor alpha (PDGFRA), TGFB1-TGFBR2/TGFBR3, and PDGFB-PDGFR complex/PDGFRA/PDGFRB/low density lipoprotein-related protein 1 (LRP1). For endothelial cells, myeloid cells promoted their proliferation through PDGFC-FLT4 and TGFB1-TGFBR3, and expressed VEGFB and VEGFA targeting FLT1 complex, FLT1, NRP1, NRP2, and KDR receptors on endothelial cells to promote angiogenesis (Figure 7C). Other cell populations, such as endothelial cells, have not been studied in depth due to their low cell numbers.
Discussion
Liver is the most common host for the metastasis of various solid malignancies, such as breast cancer, lung cancer, and many digestive system cancers, including pancreatic cancer and CRC (32,33). Nearly half of all CRC patients will develop liver metastasis in the course of the disease. Liver metastasis is the leading cause of death in CRC patients, with a mortality rate greater than 70% in patients with unresectable liver metastasis. More than 70% of patients with unresectable liver metastasis dying from liver metastasis, and approximately 30% of patients treated with hepatectomy will eventually die from liver metastasis. Currently, patients who do not undergo surgery for liver metastasis typically live less than 18 months, and 5-year survivors are rare. Patients with liver metastasis also have significantly shorter disease-specific survival compared to patients with other metastatic sites (2). Therefore, liver metastasis is a significant limiting factor for survival in CRC patients, and warrants further research and the development of clinical solutions. At present, the main treatment methods for CRC liver metastases include surgery, chemotherapy, targeted therapy, immunotherapy and interventional therapy. Medicine tolerance, high rate of local recurrence and failure to achieve no evidence of disease (NED) through surgery are the key problems to be solved. In the liver, there is a significant interaction between inflammation and cancer. During the development of liver metastasis, pro-inflammatory signaling can eliminate invading cancer cells, playing an important role in establishing a “pro-metastatic” environment conducive to invasion and colonization. In view of the poor prognosis of CRC patients with liver metastasis and the lack of effective treatment, this study used scRNA-seq to analyze the cellular heterogeneity between the primary lesion and liver metastasis of CRC patients, so as to examine the specific TME and regulatory pathways in CRC liver metastasis. This information will provide a theoretical basis for further exploration of the pathophysiological mechanisms and the development of treatment regimens for CRC patients with liver metastasis. A similar study by Zhang et al. previously identified that abnormal iron cell apoptosis mediated granulocyte death and the activation of Wnt signaling pathway promoted granulocyte migration (34). This paper focuses on the interaction between myeloid cells and other cell subsets in the TME, with different emphasis and perspective, which is helpful to fully demonstrate the characteristics of the immune microenvironment of CRC liver metastasis.
A total of 11 cell populations of epithelial cells, stromal cells, and immune cells in primary CRC and liver metastasis were detected. Among the 4 subgroups of epithelial cells, the proportion of epithelial cells 2 in liver metastasis was slightly higher than that in primary lesion, and these cells expressed elevated levels of proliferation-related genes such as MKI67, indicating that they were in a highly proliferative state. The relatively low copy number variation of epithelial cells 2 suggested that they may be in the transitional stage of malignant transformation. Further functional enrichment analysis revealed that pathways related to cell cycle and metabolism were significantly enriched in epithelial cells 2, consistent with their highly proliferative state. Epithelial cells 3 showed the greatest difference in distribution between the primary lesion and liver metastasis, almost all of which were located in the metastatic liver sample, and this subset of cells had a high degree of copy number variation. These more malignant cells occupied a higher proportion in liver metastasis than in primary tumor, which may be related to the activation of KRAS signal transduction and the enhancement of EMT in epithelial cells 3. KRAS is a downstream gene of the epidermal growth factor receptor (EGFR) signaling pathway and has the important function of regulating cell proliferation and differentiation (35). EMT is the process of cell dedifferentiation, through which epithelial cells lose epithelial phenotypes, such as cell polarity and connection with basement membrane, and acquire mesenchymal phenotypes, such as greater migration and invasion, as well as anti-apoptotic and ECM degradation capabilities (36), thereby, triggering metastasis. Therefore, epithelial cells 3 are more adaptive to the liver microenvironment, enabling survival and colonization. Compared with epithelial cells 3, epithelial cells 1 and 2 showed little difference in numbers between primary and metastatic lesions, but they both significantly upregulated Notch signaling in metastasis. The Notch signaling pathway is widely involved in cell proliferation, differentiation, and apoptosis, and plays an important role in tumorigenesis, invasion, and metastasis (37,38), as well as mediating the interaction between tumor cells and the TME. In addition, it is involved in the regulation of tumor angiogenesis, maintenance of stem cell stemness, acquisition of EMT, infiltration of immune cells, and resistance to therapy (39,40). Sustained Notch1 activity in epithelial cells will lead to increased expression of EMT/stemness-related proteins CD44, Slug, and Smad-3, leading to EMT and stem-like phenotype of CRC (41), and promoting angiogenesis in intestinal tumors (42). Furthermore, the close interaction of Notch1 with transcription factors such as Slug, Snail, and TGF-β, creates a TME that promotes CRC metastasis (43), ultimately allowing tumor cells to migrate from the primary tumor and home in distant sites. The differences in epithelial cell subsets between primary and metastatic CRC suggested that epithelial cell subsets with different biological characteristics play different roles in modeling the composition and function of the TME during liver metastasis. Epithelial cells 2 acted as “seeds” to maintain proliferation and survival by regulating cell cycle and metabolism-related pathways. Epithelial cell 3 was a cell subset further adapted to the liver environment, had stronger migration and invasion capabilities, and more actively transformed the TME to achieve immune escape and proliferation. In the process of liver metastasis, these different epithelial cell populations can coexist in different metastases or different parts of the same metastasis, and their proportions may be one of the factors affecting the progression speed and malignancy of liver metastases.
In addition to the heterogeneity of tumor cells themselves, the changes in epithelial cells from primary to metastatic tumor are also influenced by other cells in the TME. Among the non-epithelial cell populations we detected, the proportion of myeloid cells in metastasis was significantly increased, while the proportion of B cells, plasma cells, and stromal cells was significantly decreased. Myeloid cells in tumor tissues constitute a dynamic immune unit characterized by heterogeneous phenotypes and distinct functional activities. MDSCs represent a heterogeneous population of immature myeloid cells capable of modulating immune responses and play key roles in both innate and adaptive immunity. In cancer, MDSCs are abnormally generated and recruited to the TME, helping establish an immunosuppressive TME that promotes tumor escape. Liver metastases are rich in immunosuppressive cells, such as M2-like macrophages, SPP1 macrophages, neutrophils. Macrophages in primary tumors mainly express inflammatory cytokines, such as CCL3, CCL4, tumor necrosis factor (TNF) and ILIB, while macrophages in metastatic organs mainly express macrophage polarization-related genes, such as APOE and MARCO, suggesting that metabolic regulation may mediate the phenotypic and functional changes of macrophages in response to different environmental signals (44). We further compared the expression of MDSC characteristic genes between primary and metastatic lesions, and found that IL1β was highly expressed in primary tumors, while MRC1/CD206 was significantly upregulated in metastasis. Genes associated with M2 macrophages, such as SPP1, CCL18, CXCL9, CCL5, and CCL2, as well as genes related to lipid metabolism and ECM, were also highly expressed in liver metastasis. IL1β is usually a pro-inflammatory cytokine secreted by M1 macrophages. Further subgroup analysis of myeloid cells found that myeloid cells in the primary tumor were not only fewer in numbers but also dominated by monocytes and M1 macrophages with pro-inflammatory effects. In the metastatic tumor, there was a large number of TAMs with characteristics similar to M2 macrophages. Myeloid cells showed different differentiation directions in primary and metastatic tumors, which was also confirmed by pseudotime analysis. TAMs can induce the EMT/stem cell phenotype of tumor cells, and produce molecules that promote tumor cell invasion, such as secreted protein acidic and rich in cysteine (SPARC) (45) that increases the interaction of tumor cells with the ECM, and urokinase-type plasminogen activator (uPA) and cathepsins that increase tumor cell invasiveness (46), thereby destroying the ECM and promoting metastasis. Meanwhile, TAMs can also produce VEGF, TNF-α, PDGF, thymidine aldose, and other molecules, which participate in tumor angiogenesis (47). Subsequently, TAMs induce directional migration of tumor cells around blood vessels via a paracrine loop composed of tumor cell-derived colony stimulating factor (CSF)-1 and macrophage-derived epidermal growth factor (EGF) (48,49), and acts as a part of the TME of metastases (TMEM) complex (consisting of macrophages, endothelial cells, and tumor cells), increasing vascular permeability and attracting aggressive tumor cells into the circulation. We reconstructed the transcriptional regulatory network through SCENIC and analyzed the differences in transcriptional regulation levels between primary and metastatic tumors, and found that transcription factors such as ATF5, TFEC, USF2, MAF, and MAFB, which regulate M2 signature genes, have significantly increased regulatory activity in metastasis. Studies have shown that inhibition of ATF5 expression sensitizes cancer cells to anchorage-dependent death signals. The high expression of ATF5 enables CRC cells to select a suitable local environment for anchoring and evade elimination by anchorage-dependent death. In other studies, inhibition of ATF5 expression may block survival signaling in CRC cells (50). Furthermore, ATF5 is associated with tumor mitochondrial dysfunction. It is significantly upregulated in esophageal squamous cell carcinoma and associated with poor survival. ATF5 may function as a novel co-activator in the HIF1 transcriptional complex by binding to HIF1α (51). L-4/signal transducer and activator of transcription 6/TFEC/IL-4 receptor can form a positive feedback regulatory loop. IL-4 induces the expression of C/EBP homologous protein (Chop), which promotes signal transducer and activator of transcription 6 signaling to transduce TFEC expression. TFEC then transcribes IL-4 receptor alpha expression to promote M2 programming in macrophages (52). Adenomatous polyposis (APC) gene products are involved in cell cycle arrest and apoptosis, and play important roles in CRC progression. USF1 and USF2 can bind to AP2, Sp1, CAAT box, and other sites in the APC promoter to activate its transcription (53). However, increased regulatory activity of USF2 in CRC liver metastases was not shown to reduce tumor growth, possibly because USF2 transcriptionally regulates the expression of S100 calcineurin A8 (S100A8) by directly binding to the TGF-β promoter. TGF-β can promote EMT and metastasis through the USF2/S100A8 axis. USF2 is thought to be an important switch in the intracellular and extracellular S100A8 feedback loop (54). The increased regulatory activity of USF2 suggests that in liver metastases, it may contribute to the increased malignancy of metastases through TGF-β. In addition, studies have reported that microRNA (miR)-362-3p is overexpressed in colon cancer cell lines and leads to cell proliferation through cell cycle arrest, and USF2 is identified as one of the potential targets of miR-362-3p (55). The increased activity of USF2 in metastases may be an effect of miR-362-3p overexpression. C-MAF can promote IL4 secretion (56,57), which induces a primitive phenotype in TAMs and enhances tumor cell invasion through the production of pro-invasive factors such as EGF (58,59). MAFB is specifically expressed in macrophages and is a specific marker of macrophages in myeloid cells. It is specifically expressed in M2 macrophages derived from macrophage colony-stimulating factor (M-CSF), IL-10, and IL-4 (60). The upregulation of the above transcription factor activities in liver metastases of CRC reflects a marked increase in MDSCs. In addition, the gene function enrichment analysis also confirmed that the functions of angiogenesis, oxidative phosphorylation, and EMT of myeloid cells in the metastases were significantly enhanced. In the TME of CRC liver metastases, myeloid cells are dominated by MDSCs, and macrophages are polarized towards M2. With the help of the above-mentioned multiple pathways, a pro-metastatic niche is established, and cancer cells with more malignant phenotypes are successfully colonized in the metastatic organs.
TAMs are the main driver of TME immunosuppression (61). Due to the poor antigen-presenting ability of TAMs and M2, coupled with their ability to release some immunosuppressive factors, such as IL-10, to inhibit the proliferation of T cells, the adaptive immune response is suppressed. We grouped proliferating T cells and NK cells and found that the proportion of CD4+ T cells in the metastatic tumor increased significantly, while the proportion of CD8+ T cells decreased. In clinical trials of mCRC immunotherapy, patients with liver metastases benefited less from ICIs. It has been reported that the objective response rate (ORR) of CRC patients with liver metastases is significantly lower than that of patients without liver metastases, and patients with liver metastases had less infiltration of CD8+ T cells in metastatic lesions. The results of TCR sequencing also confirmed the above trend of changes in the clone expansion ratio of CD4+ and CD8+ T cells in liver metastasis, and the clone expansion level of Tregs in metastatic lesion was higher. Although a subset of CD8+ T cells had higher levels of clone expansion in the metastasis, the anti-tumor immune response exerted by these cells was not significant, probably because these CD8+ T cells were unable to recruit around and interact with surrounding tumor cells. This immune-excluded tumor may lack the normal chemokine status. We detected high level of CCL18 in metastases, most likely from TAMs. TAMs can indirectly achieve immunosuppression by releasing chemokines, which preferentially attract T cells without cytotoxic effects. CCL18 induces the release of Th2 cytokines (62). In an IL-10-dominated microenvironment, CCL-18 can drive immature dendritic cells (DCs) and T lymphocytes to locality. In the peripheral microenvironment, the aggregation of naïve T cells attracted by M2 and immature DCs may induce T cell inactivation, leading to immune tolerance (63). Immune checkpoints on T cells are currently a hot topic in tumor research and can activate immune tolerance. We compared the expression levels of immune checkpoint molecules and effectors in T cell subsets between primary and metastatic lesions, and confirmed that T cells in liver metastasis expressed higher levels of PD-1, TIM-3, LAG3, and other immune checkpoints compared to primary tumors. On the other hand, TAMs can express ligands of immune checkpoints, such as PD-L1, PD-L2, and B7 (64), thus suppressing the adaptive T cell immune response in liver metastases and promoting the recruitment of Tregs. In fact, there is an interaction between tumor cells, TAMs, and MDSCs. Not only can TAMs promote the expression of PD-L1 in tumor cells, but tumor cells can also induce PD-L1 expression in TAMs and MDSCs by regulating the COX2/mPGES1/PGE2 pathway, making them immunosuppressive (65). In addition, TAMs can also express PD-1, and the expression level of PD-1 is negatively correlated with the phagocytosis of macrophages. PD-1+ macrophages tended to exhibit an M2 phenotype, whereas PD-1-macrophages more closely resembled an M1 phenotype. Blockade of PD-1/PD-L1 can promote macrophage phagocytosis of tumor cells and inhibit tumor growth (66). Given the close relationship between TAMs and PD-1/PD-L1, targeting TAMs may help improve the response to immunotherapy in MSS mCRC.
In addition, we performed GO enrichment analysis of genes downregulated by CD4+ T cells in metastases and found that pathways related to B cell-mediated HI were significantly enriched. The immune status of malignant tumors is usually characterized by the inhibition of cell-mediated immunity (CMI) and the enhancement of HI, and this change will be beneficial to tumor progression (67). Distinctive CD4+ T cell subsets, such as Th1 or Th2, secrete special cytokines that regulate CMI and HI. For example, Th1 produce IL-2 and interferon-γ, which direct anti-tumor survival and anti-angiogenic CMI responses (68), while Th2 produce IL-4 and IL-10, which promote local HI responses, enhance tumor cells invasion, and stimulate angiogenesis (69). In the background of cancer development, B lymphocytes serve as a central component of HI and in addition to altering the local and circulating profile of cytokines, they limit antitumor immunity by inhibiting Th1 and CTL responses and supporting the tumor-promoting function of Th2 effector cells. A previous study has demonstrated that in the peripheral blood of CRC patients, the proportion of Th1 cells is significantly reduced, while the proportion of Th2 cells producing IL-4, IL-6, and/or IL-10 is significantly increased (70). Interestingly, however, in the present study, the proportion of CD4+ T cells was significantly elevated in liver metastasis, whereas B cell-mediated HI was attenuated. Low levels of antigen captured by B cells will cause antigen-exposed CD4+ T cells to enter a quiescent state. The successful entry of B cells into the germinal center (GC) requires high levels of pMHC expression. B cells enter the GC and interact with CD4+ T cells looking for appropriate signals for affinity maturation. CD4+ T cells also receive signals from B cells in the GC to differentiate themselves into quiescent memory T cells (71). Decreased proportion and function of B cells in the metastatic tumor leads to diminished antigen-presenting function. Since CD4+ T cells are essential for the development of CD8+ memory T cells, the reduction of B cells will affect the function of CD8+ memory T cells through CD4+ T cells. Although CD4+ T cells were upregulated in metastases, the antigen-exposed CD4+ T cells showed a downward trend. The role of HI in tumor immunity is not as dominant as CMI, but it also plays an important role. The differences in B cells between primary and metastatic lesions may suggest that the anti-tumor immune regulation of B cells is further weakened in the process of liver metastases. This means that, contrary to traditional views, the immune microenvironment in liver metastases of CRC is very complex, not only is CMI tolerated, but HI is also weakened. Under the “competition” of the co-attenuation of CMI and HI, the effect of the former remains dominant. Cytokines derived from evolving tumor cells, activated resident stromal cells, or infiltrating immune cells dynamically regulate tumor growth through affecting angiogenesis, cell survival, death, or differentiation.
Intercellular communication analysis showed strong interactions between myeloid cells and other cell populations, suggesting that myeloid cells play an important role in the TME. In this study, endothelial cells, epithelial cells, fibroblasts, and T and NK cells secrete chemokines such as CXCL12, CCL4, CCL5, CXCL9, and CCL14, acting on the corresponding receptors on the myeloid cells to affect the polarization of macrophages. Conversely, myeloid cells act on endothelial cells, epithelial cells, fibroblasts, T and NK cells, plasma cells, and B cells by expressing CCL4L2, CXCL16, CXCL9, CCL5, CCL4, CCL3, and CXCL8, inducing the orientation of these cells. In the TME, cancer cells destabilize the normal environment of inflammatory cytokines and chemokines. By secreting chemokines, cancer cells recruit T cells, monocytes, myeloid cells, fibroblasts, and mesenchymal stem cells (MSCs), which are then individually transformed into Tregs (72), M2-TAMs (73), MDSCs (74), CAFs (75), and tumor-associated adipocytes (76), forming a tumor-protective microenvironment composed of anergic or exhausted T cells and NKs. CXCL12 is a homeostatic chemokine and the CXCL12/CXCR4 axis is involved in tumor progression, angiogenesis, metastases, and survival. Several exosomal miRNAs in CRC cells can be upregulated by activating the CXCL12/CXCR4 axis, and they induce M2 polarization of macrophages by activating the PI3K/Akt signaling pathway to regulate PTEN. Finally, M2-polarized macrophages promote cancer metastasis by enhancing EMT and secreting VEGF (77). CXCL9, induced by IFN-γ, mainly mediates lymphocyte infiltration to focal sites and inhibits tumor growth. Studies have shown that the CXCL9/CXCR3 axis can affect TAMs polarization (78). The CCL5/CCR5 axis is involved in immunosuppressive TAM polarization (79), and monocytes recruited by CCL5 are converted to immunosuppressive M2 (80). CCL5 recruits T cells and CCR5+ Tregs, the latter of which is inhibitory to tumoricidal lymphocytes (19). Fibroblasts and MSCs are recruited by CCR5 ligands and induce the secretion of CCL5 to become CAFs (81,82), which not only support cancer cell proliferation, migration, metastases, and EMT, but also recruit monocytes and Tregs to suppress effector T cells (83). In the bone marrow, hematopoietic cells overexpressing CCL5 become MDSCs, serving as a heterogeneous population of myeloid cells that limit immune responses to tumors (84). Moreover, we found that myeloid cell-derived CCL4 mainly acts on tumor cell populations. CCL4, also known as macrophage inflammatory protein (MIP)-1β, interacts with its specific receptor CCR5 and cooperates with other related chemokines such as CCL3 and CCL5, to exert multiple effects on various immune and non-immune cells. CCL4 can promote tumor progression by recruiting Tregs and tumor-promoting macrophages, and enhance the tumor-promoting capacity of other resident cells in the TME, such as fibroblasts and endothelial cells (85).
Moreover, epithelial cells, endothelial cells, and fibroblasts act on myeloid cells through growth factors such as TGFB1, PDGFB, IGF2, PGF, HGF, and HBEGF. Myeloid cells express HBEGF, which acts on almost all cells except endothelial cells, promoting their proliferation. CD44 is a widely distributed transmembrane glycoprotein, and studies have shown that heparan sulfate (HS)-modified CD44 isoforms can bind to the HS-binding growth factor HBEGF (86). Activated CD44 further activates relevant intracellular signaling pathways to induce cell proliferation, increase cell survival, modulate cytoskeletal changes, and enhance cell motility (87). For endothelial cells, myeloid cells promote their proliferation through PDGFC-FLT4 and TGFB1-TGFBR3, and by expressing VEGFA and VEGFB, they act on the corresponding receptors on endothelial cells to promote angiogenesis. All these results suggest that myeloid cells not only play a key regulatory role in suppressing anti-tumor immune responses but also interact with various types of cells in the TME, mediating complex effects that promote tumor cell proliferation, angiogenesis, invasion, and metastasis. Due to the small number of other cell populations, we did not conduct in-depth research. In the future, analysis of different samples should be conducted to investigate the functional mechanism of endothelial cells and other components in the TME.
Conclusions
Liver metastasis of CRC is a complex biological process involving multiple factors and steps. By performing scRNA-seq, we demonstrated that liver metastasis is significantly immunosuppressed relative to primary CRC. In liver metastases, myeloid cells are dominated by MDSCs, macrophages are polarized towards the M2 state, and T cells are in a state of exhaustion. Myeloid cells interact with other cell populations in the TME, by influencing angiogenesis, tumor cell invasiveness, and EMT, and they establish a pro-metastatic niche that helps CRC cells colonize and grow in the liver. MSS mCRC is naturally resistant to ICIs, and TAM-targeted therapy may be a promising method to resolve the dilemma of MSS mCRC immunotherapy.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 82002479), the Natural Science Foundation of Jiangsu Province (No. BK20200179), the Basic Research Project of Changzhou Science and Technology Bureau (No. CJ20200116), the Young Talent Development Plan of Changzhou Health Commission (No. CZQM2020006), the Young Scientific and Technological Talent Support Project of Changzhou (2021), the Special Plan for Bring in Foreign Talents (No. CQ20214031), Changzhou “14th Five-Year Plan” High-level Health Personnel Training Project (No. KY20221354 and No. KY20221355), and the Science and Technology Program for Youth Talent of Changzhou Health Commission (No. QN202015).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5270/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5270/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5270/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Ethics Committee of The Third Affiliated Hospital of Soochow University (No. 2022-science-158). As this was a retrospective study, only tumor tissue samples were used, and the patient could not be contacted, the ethics committee approved this study to be exempt from the informed consent process.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. [Crossref] [PubMed]
- Stewart CL, Warner S, Ito K, et al. Cytoreduction for colorectal metastases: liver, lung, peritoneum, lymph nodes, bone, brain. When does it palliate, prolong survival, and potentially cure? Curr Probl Surg 2018;55:330-79. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Fukumura D, Yuan F, Monsky WL, et al. Effect of host microenvironment on the microcirculation of human colon adenocarcinoma. Am J Pathol 1997;151:679-88. [PubMed]
- Denève E, Riethdorf S, Ramos J, et al. Capture of viable circulating tumor cells in the liver of colorectal cancer patients. Clin Chem 2013;59:1384-92. [Crossref] [PubMed]
- Rahbari NN, Bork U, Kircher A, et al. Compartmental differences of circulating tumor cells in colorectal cancer. Ann Surg Oncol 2012;19:2195-202. [Crossref] [PubMed]
- Clark AM, Ma B, Taylor DL, et al. Liver metastases: Microenvironments and ex-vivo models. Exp Biol Med (Maywood) 2016;241:1639-52. [Crossref] [PubMed]
- Lee JW, Stone ML, Porrett PM, et al. Hepatocytes direct the formation of a pro-metastatic niche in the liver. Nature 2019;567:249-52. [Crossref] [PubMed]
- Brodt P. Role of the Microenvironment in Liver Metastasis: From Pre- to Prometastatic Niches. Clin Cancer Res 2016;22:5971-82. [Crossref] [PubMed]
- Milette S, Sicklick JK, Lowy AM, et al. Molecular Pathways: Targeting the Microenvironment of Liver Metastases. Clin Cancer Res 2017;23:6390-9. [Crossref] [PubMed]
- Keirsse J, Van Damme H, Geeraerts X, et al. The role of hepatic macrophages in liver metastasis. Cell Immunol 2018;330:202-15. [Crossref] [PubMed]
- Cortese N, Soldani C, Franceschini B, et al. Macrophages in Colorectal Cancer Liver Metastases. Cancers (Basel) 2019;11:633. [Crossref] [PubMed]
- Vogel DY, Glim JE, Stavenuiter AW, et al. Human macrophage polarization in vitro: maturation and activation methods compared. Immunobiology 2014;219:695-703. [Crossref] [PubMed]
- Lee JJ, Chu E. Recent Advances in the Clinical Development of Immune Checkpoint Blockade Therapy for Mismatch Repair Proficient (pMMR)/non-MSI-H Metastatic Colorectal Cancer. Clin Colorectal Cancer 2018;17:258-73. [Crossref] [PubMed]
- Fukuoka S, Hara H, Takahashi N, et al. Regorafenib Plus Nivolumab in Patients With Advanced Gastric or Colorectal Cancer: An Open-Label, Dose-Escalation, and Dose-Expansion Phase Ib Trial (REGONIVO, EPOC1603). J Clin Oncol 2020;38:2053-61. [Crossref] [PubMed]
- Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014;344:1396-401. [Crossref] [PubMed]
- Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol 2018;18:35-45. [Crossref] [PubMed]
- Huang S, Tan X, Huang Z, et al. microRNA biomarkers in colorectal cancer liver metastasis. J Cancer 2018;9:3867-73. [Crossref] [PubMed]
- Mantovani A, Marchesi F, Malesci A, et al. Tumour-associated macrophages as treatment targets in oncology. Nat Rev Clin Oncol 2017;14:399-416. [Crossref] [PubMed]
- Hanley CJ, Mellone M, Ford K, et al. Targeting the Myofibroblastic Cancer-Associated Fibroblast Phenotype Through Inhibition of NOX4. J Natl Cancer Inst 2018;110:109-120. [Crossref] [PubMed]
- Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 2018;24:541-50. [Crossref] [PubMed]
- Galon J, Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat Rev Drug Discov 2019;18:197-218. [Crossref] [PubMed]
- Junttila MR, de Sauvage FJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 2013;501:346-54. [Crossref] [PubMed]
- Bindea G, Mlecnik B, Tosolini M, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 2013;39:782-95. [Crossref] [PubMed]
- Mlecnik B, Bindea G, Angell HK, et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity 2016;44:698-711. [Crossref] [PubMed]
- Zhang L, Zhang Z. Recharacterizing Tumor-Infiltrating Lymphocytes by Single-Cell RNA Sequencing. Cancer Immunol Res 2019;7:1040-6. [Crossref] [PubMed]
- Chen Z, Zhou L, Liu L, et al. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat Commun 2020;11:5077. [Crossref] [PubMed]
- Bi K, He MX, Bakouny Z, et al. Tumor and immune reprogramming during immunotherapy in advanced renal cell carcinoma. Cancer Cell 2021;39:649-661.e5. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Aibar S, González-Blas CB, Moerman T, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods 2017;14:1083-6. [Crossref] [PubMed]
- Sakai H, Iwatani Y, Katayama Y. Analysis of lymphocyte subpopulation and subsets in patients with idiopathic cardiomyopathy. Rinsho Byori 1988;36:705-9. [PubMed]
- Obenauf AC, Massagué J. Surviving at a Distance: Organ-Specific Metastasis. Trends Cancer 2015;1:76-91. [Crossref] [PubMed]
- Hess KR, Varadhachary GR, Taylor SH, et al. Metastatic patterns in adenocarcinoma. Cancer 2006;106:1624-33. [Crossref] [PubMed]
- Zhang Y, Song J, Zhao Z, et al. Single-cell transcriptome analysis reveals tumor immune microenvironment heterogenicity and granulocytes enrichment in colorectal cancer liver metastases. Cancer Lett 2020;470:84-94. [Crossref] [PubMed]
- Seger R, Krebs EG. The MAPK signaling cascade. FASEB J 1995;9:726-35. [Crossref] [PubMed]
- Steinestel K, Eder S, Schrader AJ, et al. Clinical significance of epithelial-mesenchymal transition. Clin Transl Med 2014;3:17. [Crossref] [PubMed]
- Meurette O, Mehlen P. Notch Signaling in the Tumor Microenvironment. Cancer Cell 2018;34:536-48. [Crossref] [PubMed]
- Allenspach EJ, Maillard I, Aster JC, et al. Notch signaling in cancer. Cancer Biol Ther 2002;1:466-76. [Crossref] [PubMed]
- Sethi N, Dai X, Winter CG, et al. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell 2011;19:192-205. [Crossref] [PubMed]
- Timmerman LA, Grego-Bessa J, Raya A, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev 2004;18:99-115. [Crossref] [PubMed]
- Fender AW, Nutter JM, Fitzgerald TL, et al. Notch-1 promotes stemness and epithelial to mesenchymal transition in colorectal cancer. J Cell Biochem 2015;116:2517-27. [Crossref] [PubMed]
- Rodilla V, Villanueva A, Obrador-Hevia A, et al. Jagged1 is the pathological link between Wnt and Notch pathways in colorectal cancer. Proc Natl Acad Sci U S A 2009;106:6315-20. [Crossref] [PubMed]
- Jackstadt R, van Hooff SR, Leach JD, et al. Epithelial NOTCH Signaling Rewires the Tumor Microenvironment of Colorectal Cancer to Drive Poor-Prognosis Subtypes and Metastasis. Cancer Cell 2019;36:319-336.e7. [Crossref] [PubMed]
- Wu Y, Yang S, Ma J, et al. Spatiotemporal Immune Landscape of Colorectal Cancer Liver Metastasis at Single-Cell Level. Cancer Discov 2022;12:134-53. [Crossref] [PubMed]
- Sangaletti S, Di Carlo E, Gariboldi S, et al. Macrophage-derived SPARC bridges tumor cell-extracellular matrix interactions toward metastasis. Cancer Res 2008;68:9050-9. [Crossref] [PubMed]
- Hildenbrand R, Wolf G, Böhme B, et al. Urokinase plasminogen activator receptor (CD87) expression of tumor-associated macrophages in ductal carcinoma in situ, breast cancer, and resident macrophages of normal breast tissue. J Leukoc Biol 1999;66:40-9. [Crossref] [PubMed]
- Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell 2010;141:39-51. [Crossref] [PubMed]
- Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell 2006;124:263-6. [Crossref] [PubMed]
- Wyckoff JB, Wang Y, Lin EY, et al. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res 2007;67:2649-56. [Crossref] [PubMed]
- Piran M, Sepahi N, Moattari A, et al. Systems Biomedicine of Primary and Metastatic Colorectal Cancer Reveals Potential Therapeutic Targets. Front Oncol 2021;11:597536. [Crossref] [PubMed]
- He F, Xiao H, Cai Y, et al. ATF5 and HIF1α cooperatively activate HIF1 signaling pathway in esophageal cancer. Cell Commun Signal 2021;19:53. [Crossref] [PubMed]
- Wang Y, Zhu J, Zhang L, et al. Role of C/EBP homologous protein and endoplasmic reticulum stress in asthma exacerbation by regulating the IL-4/signal transducer and activator of transcription 6/transcription factor EC/IL-4 receptor α positive feedback loop in M2 macrophages. J Allergy Clin Immunol 2017;140:1550-61.e8. Correction in J Allergy Clin Immunol 2022;150:491. [Crossref] [PubMed]
- Jaiswal AS, Narayan S. Upstream stimulating factor-1 (USF1) and USF2 bind to and activate the promoter of the adenomatous polyposis coli (APC) tumor suppressor gene. J Cell Biochem 2001;81:262-77. [Crossref] [PubMed]
- Li S, Zhang J, Qian S, et al. S100A8 promotes epithelial-mesenchymal transition and metastasis under TGF-β/USF2 axis in colorectal cancer. Cancer Commun (Lond) 2021;41:154-70. [Crossref] [PubMed]
- Gottschalk A. Commentary on "Patient-reported outcomes after 3-dimensional conformal, intensity-modulated, or proton beam radiotherapy for localized prostate cancer." Gray PJ, Paly JJ, Yeap BY, Sanda MG, Sandler HM, Michalski JM, Talcott JA, Coen JJ, Hamstra DA, Shipley WU, Hahn SM, Zietman AL, Bekelman JE, Efstathiou JA. Harvard Radiation Oncology Program, Boston, MA.: Cancer 2013;119(9):1729-35. doi:
10.1002/cncr.27956 . [Epub 2013 Feb 22]. Urol Oncol 2014;32:373-74.10.1002/cncr.27956 - Kwon SJ, Crespo-Barreto J, Zhang W, et al. KLF13 cooperates with c-Maf to regulate IL-4 expression in CD4+ T cells. J Immunol 2014;192:5703-9. [Crossref] [PubMed]
- Lai CY, Lin SY, Wu CK, et al. Tyrosine phosphorylation of c-Maf enhances the expression of IL-4 gene. J Immunol 2012;189:1545-50. [Crossref] [PubMed]
- Shapouri-Moghaddam A, Mohammadian S, Vazini H, et al. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol 2018;233:6425-40. [Crossref] [PubMed]
- DeNardo DG, Barreto JB, Andreu P, et al. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 2009;16:91-102. [Crossref] [PubMed]
- Daassi D, Hamada M, Jeon H, et al. Differential expression patterns of MafB and c-Maf in macrophages in vivo and in vitro. Biochem Biophys Res Commun 2016;473:118-24. [Crossref] [PubMed]
- Yang L, Zhang Y. Tumor-associated macrophages: from basic research to clinical application. J Hematol Oncol 2017;10:58. [Crossref] [PubMed]
- Wang Q, Tang Y, Yu H, et al. CCL18 from tumor-cells promotes epithelial ovarian cancer metastasis via mTOR signaling pathway. Mol Carcinog 2016;55:1688-99. [Crossref] [PubMed]
- Goswami KK, Ghosh T, Ghosh S, et al. Tumor promoting role of anti-tumor macrophages in tumor microenvironment. Cell Immunol 2017;316:1-10. [Crossref] [PubMed]
- Mantovani A, Marchesi F, Jaillon S, et al. Tumor-associated myeloid cells: diversity and therapeutic targeting. Cell Mol Immunol 2021;18:566-78. [Crossref] [PubMed]
- Prima V, Kaliberova LN, Kaliberov S, et al. COX2/mPGES1/PGE2 pathway regulates PD-L1 expression in tumor-associated macrophages and myeloid-derived suppressor cells. Proc Natl Acad Sci U S A 2017;114:1117-22. [Crossref] [PubMed]
- Gordon SR, Maute RL, Dulken BW, et al. PD-1 expression by tumour-associated macrophages inhibits phagocytosis and tumour immunity. Nature 2017;545:495-9. [Crossref] [PubMed]
- Dalgleish AG, O'Byrne KJ. Chronic immune activation and inflammation in the pathogenesis of AIDS and cancer. Adv Cancer Res 2002;84:231-76. [Crossref] [PubMed]
- Qin Z, Blankenstein T. CD4+ T cell--mediated tumor rejection involves inhibition of angiogenesis that is dependent on IFN gamma receptor expression by nonhematopoietic cells. Immunity 2000;12:677-86. [Crossref] [PubMed]
- Sakamoto T, Saito H, Tatebe S, et al. Interleukin-10 expression significantly correlates with minor CD8+ T-cell infiltration and high microvessel density in patients with gastric cancer. Int J Cancer 2006;118:1909-14. [Crossref] [PubMed]
- Kanazawa M, Yoshihara K, Abe H, et al. Effects of PSK on T and dendritic cells differentiation in gastric or colorectal cancer patients. Anticancer Res 2005;25:443-9. [PubMed]
- Welsh RA, Song N, Sadegh-Nasseri S. How Does B Cell Antigen Presentation Affect Memory CD4 T Cell Differentiation and Longevity? Front Immunol 2021;12:677036. [Crossref] [PubMed]
- Chang LY, Lin YC, Mahalingam J, et al. Tumor-derived chemokine CCL5 enhances TGF-β-mediated killing of CD8(+) T cells in colon cancer by T-regulatory cells. Cancer Res 2012;72:1092-102. [Crossref] [PubMed]
- Cook J, Hagemann T. Tumour-associated macrophages and cancer. Curr Opin Pharmacol 2013;13:595-601. [Crossref] [PubMed]
- Bronte V, Brandau S, Chen SH, et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat Commun 2016;7:12150. [Crossref] [PubMed]
- Yang X, Hou J, Han Z, et al. One cell, multiple roles: contribution of mesenchymal stem cells to tumor development in tumor microenvironment. Cell Biosci 2013;3:5. [Crossref] [PubMed]
- Wu Q, Li B, Li Z, et al. Cancer-associated adipocytes: key players in breast cancer progression. J Hematol Oncol 2019;12:95. [Crossref] [PubMed]
- Wang D, Wang X, Si M, et al. Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett 2020;474:36-52. [Crossref] [PubMed]
- Tokunaga R, Zhang W, Naseem M, et al. CXCL9, CXCL10, CXCL11/CXCR3 axis for immune activation - A target for novel cancer therapy. Cancer Treat Rev 2018;63:40-7. [Crossref] [PubMed]
- Halama N, Zoernig I, Berthel A, et al. Tumoral Immune Cell Exploitation in Colorectal Cancer Metastases Can Be Targeted Effectively by Anti-CCR5 Therapy in Cancer Patients. Cancer Cell 2016;29:587-601. [Crossref] [PubMed]
- Casagrande N, Borghese C, Visser L, et al. CCR5 antagonism by maraviroc inhibits Hodgkin lymphoma microenvironment interactions and xenograft growth. Haematologica 2019;104:564-75. [Crossref] [PubMed]
- Bankov K, Döring C, Ustaszewski A, et al. Fibroblasts in Nodular Sclerosing Classical Hodgkin Lymphoma Are Defined by a Specific Phenotype and Protect Tumor Cells from Brentuximab-Vedotin Induced Injury. Cancers (Basel) 2019;11:1687. [Crossref] [PubMed]
- Barcellos-de-Souza P, Comito G, Pons-Segura C, et al. Mesenchymal Stem Cells are Recruited and Activated into Carcinoma-Associated Fibroblasts by Prostate Cancer Microenvironment-Derived TGF-β1. Stem Cells 2016;34:2536-47. [Crossref] [PubMed]
- Ansems M, Span PN. The tumor microenvironment and radiotherapy response; a central role for cancer-associated fibroblasts. Clin Transl Radiat Oncol 2020;22:90-7. [Crossref] [PubMed]
- Ban Y, Mai J, Li X, et al. Targeting Autocrine CCL5-CCR5 Axis Reprograms Immunosuppressive Myeloid Cells and Reinvigorates Antitumor Immunity. Cancer Res 2017;77:2857-68. [Crossref] [PubMed]
- Mukaida N, Sasaki SI, Baba T. CCL4 Signaling in the Tumor Microenvironment. Adv Exp Med Biol 2020;1231:23-32. [Crossref] [PubMed]
- Bennett KL, Jackson DG, Simon JC, et al. CD44 isoforms containing exon V3 are responsible for the presentation of heparin-binding growth factor. J Cell Biol 1995;128:687-98. [Crossref] [PubMed]
- Chen C, Zhao S, Karnad A, et al. The biology and role of CD44 in cancer progression: therapeutic implications. J Hematol Oncol 2018;11:64. [Crossref] [PubMed]
(English Language Editor: J. Teoh)


