
Silencing the expression of lncRNA SNHG15 may be a novel therapeutic approach in human breast cancer through regulating miR-345-5p
Introduction
Breast cancer is a substantial health burden for women globally, and despite significant breakthroughs in early detection and treatment regimens (1), metastasis of breast cancer can result in high mortality and poor prognosis (2,3). Breast cancer is the result of a multi-step process that can be triggered by aberrant gene expression or epigenetic alteration (4,5). There has been much research in recent decades investigating the molecular pathways that govern the genesis of breast cancer. Long noncoding RNAs (lncRNA) appear to have a key role in the genesis and progression of breast cancer (6). Furthermore, dysregulated lncRNAs have been linked to cisplatin resistance in a variety of malignancies (7-9). However, the precise roles of lncRNAs in breast cancer remain to be fully elucidated.
The lncRNAs are noncoding RNAs that have more than 200 nucleotides, are unable to produce proteins, and behave as microRNA (miRNA) sponges to regulate their targets (10,11). The expression of lncRNA short nucleolar RNA host gene 15 (SNHG15) was found to be elevated in a range of malignancies, including breast cancer, and overexpression of lnc-SNHG15 has been linked to increased proliferation and metastases of tumor cells (12-14). Furthermore, lnc-SNHG15 has been shown to be an important predictor of chemoresistance (15). However, the functional relevance of lnc-SNHG15 in cisplatin resistance of breast cancer is not fully understood.
The miRNAs are short non-coding RNAs (usually 18–22 nucleotides) which play an important role in gene expression and regulation after transcription (16). MiRNAs interact with a number of target coding mRNAs, causing the degradation or translation inhibition of the mRNAs. A review article has summarized that miRNAs participate in a variety of biological processes, including cancer formation (17). Increasingly, reports have suggested that miRNAs may act as oncogenes or tumor suppressors in cancers. Recently, a study found that the expression of miR-345-5p was much higher in the serum of prostate cancer patients than in the serum of non-tumor patients, and that miR-345-5p enhanced prostate cancer cell proliferation and metastasis (18). Another study found that the expression of miR-345-5p was suppressed in gastric cancer and that miR-345-5p might block epithelial-mesenchymal transition, thereby preventing cancer cells from spreading (19). Furthermore, in non-small cell lung cancer, miR-345-5p has been shown to be downregulated and associated with patient prognosis. These findings suggested that miR-345-5p has numerous functions in human malignancies (20). However, it is uncertain if miR-345-5p plays a role in the pathophysiology of breast cancer.
The zebrafish model has been applied to explore the oncogenic roles of lncRNAs LINC00152 and THOR in the advancement of certain malignancies (21,22). In addition, earlier research has shown that human colorectal cancer cell lines may be successfully transplanted into zebrafish to test their proliferation and migratory capabilities (23). Based on these findings, the zebrafish xenograft model may be useful for investigating the involvement of lncRNAs in breast cancer.
This current study examined the expression pattern and the underlying molecular mechanisms of lnc-SNHG15 in cisplatin resistant breast cancer. Lnc-SNHG15 expression was upregulated in the breast cancer cell line MDA-MB-231 and in the zebrafish xenograft model. Knockdown of lnc-SNHG15 expression rendered the breast cancer cells more sensitive to cisplatin, thereby, suppressing the tumor. Lnc-SNHG15 knockdown also increased miR-345-5p expression in breast cancer cells, rendering them more sensitive to cisplatin. This novel lnc-SNHG15/miR-345-5p regulatory pathway may be a potential therapeutic target in cisplatin resistant breast cancer. We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5275/rc).
Methods
Cell culture
The human breast cancer cell line MDA-MB-231 and the human breast epithelial cell line MCF-10A were obtained from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). The MDA-MB-231 cells were grown in DMEM (GIBCO, C11995500BT), supplemented with 10% fetal bovine serum (FBS; ScienCell, 0500). The MCF-10A cells were cultured in DMEM (GIBCO, C11995500BT), supplemented with 5% horse serum (Procell, 164215). Both media contained 100 U/mL penicillin and 100 mg/mL streptomycin (Gibco, 15140122). All cell cultures were maintained at 37 ℃ and 5% CO2.
RNA isolation and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was extracted from cells using Trizol reagent (ambion, 15596026) and 1 g RNA from each sample was reverse transcribed to cDNA using random primers and the Hifair® II 1st Strand cDNA Synthesis Kit (gDNA digester plus) (YEASEN, 11121ES60), as per the manufacturer’s protocols. The miR-345-5p and miR-627 were reverse transcribed to cDNA using the following primers, GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACGAGCCC and GTTGGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACACCCAA, respectively. Real-time PCR was performed using Hieff® qPCR SYBR Green Master Mix (High Rox Plus) (YEASEN, 11203ES08), according to the instructions of the manufacturer. The expression of SNHG15 was normalized to that of glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and the expression of miR-345-5p and miR-623 were normalized to that of U6. The comparative cycle threshold (CT) (2−ΔΔCT) method was used to calculate the relative gene expression levels. The primer sequences for real-time PCR are presented in Table 1.
Table 1
| Gene | Forward primer (5'-3') | Reverse primer (5'-3') |
|---|---|---|
| SNHG15 | CCAGTGGCTTCACTCTGTGT | GGGACCTGACCTGAGAGAAGAT |
| GAPDH | GGGAGCCAAAAGGGTCAT | GAGTCCTTCCACGATACCAA |
| MiR-345-5p | GTTGCTGACTCCTAGTCCA | GTGCAGGGTCCGAGGT |
| MiR-623 | TTGATCCCTTGCAGGGGC | GTGCAGGGTCCGAGGT |
| U6 | GCTTCGGCAGCACATATACTAAAAT | CGCTTCACGAATTTGCGTGTCAT |
PCR, polymerase chain reaction.
RNA interference
MDA-MB-231 cells were seeded in 6-well plates until they reached 70–80% confluency and transfected with the specific siRNAs (50 nM) or miR inhibitor (100 nM) using Lipofectamine 2000 (Invitrogen, 11668019). After 24 hours, the cells were harvested, and the transfection efficiency was assessed by qRT-PCR. The negative control siRNA (NC) and 3 SNHG15 siRNAs (si1-SNHG15, si2-SNHG15, si3-SNHG15), as well as the miR-335-5p inhibitor, were obtained from General Biol (Chuzhou, China). The si1-SNHG15, si2-SNHG15, si3-SNHG15 sequences are listed below in Table 2.
Table 2
| Si-RNAs | Sequence |
|---|---|
| si1-SNHG15 | 5'-GGACCTGACCTGAGAGAAG-3' |
| si2-SNHG15 | 5'-CCAAGATGCTGGAGCTAGA-3' |
| si3-SNHG15 | 5'-AGTGAGGAGTGGAGCTGAA-3' |
| Negative control siRNA | 5'-TTCTCCGAACGTGTCACGT-3' |
| miR-345-5p inhibitor | 5'-GAGCCCTGGACTAGGAGTCAGC-3' |
siRNA, small interfering RNA.
Cell proliferation assay
Cell proliferation was assessed using the Cell Counting Kit-8 (CCK-8, DOJINDO, ck04). The transfected cells were collected and seeded into 96-well plates at 2×103 cells per well in 100 µL of DMEM supplemented with 10% FBS. The CCK-8 reagent (10 µL) was added to each well and cultured for 2 hours at 37 ℃. Cell proliferation was measured using a microplate reader (BioTek Elx800, USA) at 450 nm, at 0, 24, 48, and 96 hours, and the cell proliferation rate was calculated.
Colony formation assay
The transfected cells were counted and seeded into a 6-well plate at 600 cells per well and maintained for 14 days at 37 ℃. The cells were cultured in 2 mL medium supplemented with 10% FBS, 100 mg/mL streptomycin, and 100 U/mL penicillin, and the medium was replaced every 3 days. Two weeks later, cells in the plate were washed twice using Phosphate buffered saline (PBS), and fixed with methanol (Nanjing Reagent, C0690110225) for 30 minutes. The samples were then stained with 0.1% crystal violet (Solarbio, C8470) solution for 30 minutes. The total number of cell colonies was counted.
Transwell assays
MDA-MB-231 cells were transfected with SNHG15 siRNA and the negative control siRNA. After 24 hours of transfection, the cells were harvested and counted. The cells were resuspended in serum-free culture medium and [4–5]×104 cells were plated onto the upper chamber of the transwell (8-mm chamber inserts; Corning, 3422) in 24-well plates. Meanwhile, 800 µL DMEM supplemented with 10% FBS was placed into the lower chambers of the 24-well plates. After 24 hours, the inserted chambers were fixed with methanol for 20 minutes and stained with 0.1% crystal violet (solarbio, C8470) for 30 minutes. Cotton swabs were used to remove the cells on the upper surface of the membrane, and the migrated cells on the bottom surface of the membrane were photographed under an inverted microscope.
Zebrafish xenograft injection
The transfected cells were washed with Hank’s balanced salt solution (HBSS; Gibco, C14175500BT) 3 times and digested with 0.25% trypsin. The cells were then collected and resuspended in 1 mL HBSS. CM-DiI (1 µL; Invitrogen, C7000) was added to the suspension and incubated for 5 minutes at 37 ℃, following by 15 minutes at 4 ℃. Subsequently, the cells were centrifugation and washed 3 times with HBSS. Before injection, 2-day-postfertilization (dpf) zebrafish larvae were fixed using 1.2% low-melting gel (Promega, USA). Approximately 400 cells labeled with CM-DiI were injected into the perivitelline space (PVS) of the larvae using a microinjector (Picosprizer III, USA). The xenografts were cultured in a 34 ℃ incubator. On the second day after injection, the zebrafish larvae that successfully transplanted with similar sizes were collected and cultured at 34 ℃ until the end of the experiments. All zebrafish experiments were approved by the Institutional Animal Care and Use Committee of Taizhou Hospital of Zhejiang Province Affiliated to Wenzhou Medical University (Linhai, China), in compliance with institutional guidelines for the care and use of animals. The project license No. EZY-2020038.
In vivo imaging and data analysis
On the fourth day after transplantation, the zebrafish larvae were fixed with low melting point agarose gel for imaging under a confocal microscope using the 20× water-immersion objective (Fluoview 1000, Olympus, Japan) or the stereomicroscope (MVX10, Olympus, Japan), with spatial resolution of the images being 1,024×1,024 pixels (Fluoview 1000) or 1,600×1,200 (MVX10), respectively.
Statistical analysis
All the data are expressed as mean ± standard deviation (SD). P values <0.05 were considered statistically significant. Significance testing was two-tailed in a two-group comparison. Either one-way analysis of variance (ANOVA) followed by Dunnett’s post hoc test or two-way ANOVA followed by Bonferroni post hoc test was performed to determine the difference among three or more groups. Statistical analysis was conducted using GraphPad Prism software (version 8.0).
Results
The expression of lnc-SNHG15 is upregulated in human MDA-MB-231 breast cancer cells
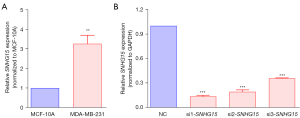
To investigate the role of lnc-SNHG15 in breast cancer, its expression in cancerous and non-cancerous human breast cell lines was assessed. The expression of lncRNA SNHG15 was significantly elevated in MDA-MB-231 human breast cancer cells compared to MCF-10A human normal breast epithelial cells, indicating that overexpression of lnc-SNHG15 may play a key function in human breast cancer (Figure 1A). To further confirm the role of SNHG15, MDA-MB-231 cells were transfected with SNHG15 siRNA (si1-SNHG15, si2-SNHG15, si3-SNHG15). Functionally, siRNA blocks gene expression by targeting and disintegrating the expanding mRNA (both exogenous and endogenous) (24). The introduction of si1-RNA, si2-RNA, and si3-RNA dramatically decreased the SNHG15 expression in MDA-MB-231 cells (Figure 1B), with si1-RNA showing the most potent inhibitory effect on SNHG15 expression.
Knockdown of lnc-SNHG15 expression downregulates the cell viability and proliferation of MDA-MB-231 cells in vitro
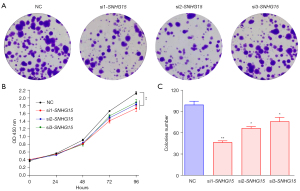
In MDA-MB-231 cells, silencing the lnc-SNHG15 gene with si1-RNA, si2-RNA, si3-RNA lowered cell viability (Figure 2A,2B) compared to the negative control siRNA, suggesting that lnc-SNHG15 knockdown inhibited cell proliferation in MDA-MB-231. In addition, the colony formation assay demonstrated that transfection with si1-SNHG15, si2-SNHG15, and si3-SNHG15 significantly decreased the number of colonies in MDA-MB-231 cells, suggestive of a low proliferation rate, compared to the negative control siRNA (Figure 2C). These results suggested that lnc-SNHG15 may increase cell proliferation and suppress apoptotic cell death, which may lead to breast cancer cell growth.
Knockdown of lnc-SNHG15 expression inhibits the migration of MDA-MB-231 cells in vitro
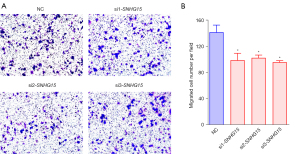
The ability of live cells to migrate is essential for normal development, immunological response, and disease processes, including cancer metastasis and inflammation (25). Hence, we investigated whether SNHG15 knockdown limits the migration of breast cancer cells. Transwell assays showed that SNGH15 knockdown downregulated migration compared to cells transfected with the negative control siRNA, suggesting that SNHG15 knockdown may be an effective approach to suppress breast cancer cells (Figure 3A,3B). Among the 3 siRNAs, si1-SNHG15 produced the most efficient lnc-SNHG15 knockdown results and was used for all subsequent experiments.
Knockdown of lnc-SNHG15 suppresses MDA-MB-231 cell growth and migration in the zebrafish xenograft model
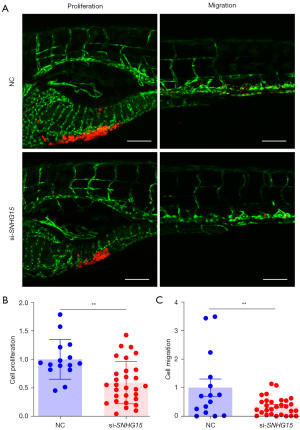
Zebrafish xenografts provide a number of distinct benefits for cancer investigations, including the optical transparency of the embryo, which allows for simple in vivo monitoring, and the ability to examine migration and proliferation concurrently at 96 hours (23,26,27). In this study, the zebrafish xenograft model was used to evaluate whether knockdown of lnc-SNHG15 by siRNAs affects the function of breast cancer. MDA-MB-231 cells transfected with si-SNHG15 and stained with a fluorescent dye (CM-Dil) were transplanted into zebrafish embryos. At 4 dpi, the yolk and trunk of the larvae were imaged and the CM-Dil positive signals were quantified. The tumor area of the yolk represents the cell proliferation, and trunk represents cell migration. The silenced-SNHG15 group showed obviously smaller quantities of fluorescent signals compared to the negative control siRNA group (Figure 4A), suggesting that cell migration and proliferation were decreased after silencing SNHG15 in MDA-MB-231 cells (Figure 4B,4C).
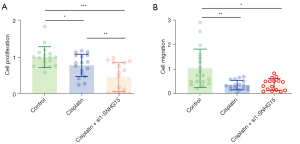
Cisplatin treatment combined with lnc-SNHG15 knockdown synergistically decreases cell growth and migration in the zebrafish xenograft
Cisplatin is a chemotherapy drug often used to treat malignancies. It works by interfering with DNA replication and transcription, and thus, rapidly replicating tumor cells are more sensitive to cisplatin than normal cells (28). Cisplatin dramatically reduced breast cancer cell growth and migration in the zebrafish xenograft model compared to controls (Figure 5A,5B). Furthermore, after silencing the SNHG15 gene using si1-RNA, the effect of cisplatin was enhanced, suggesting a synergistic effect between SNHG15 silencing and cisplatin.
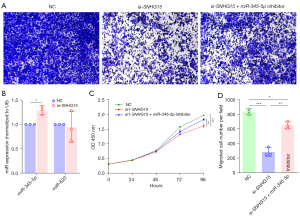
Lnc-SNHG15 negatively regulates the expression of miR-345-5p in MDA-MB-231 cells
The miR-345-5p and miR-623 genes are responsible for the suppression of cell proliferation and migration in cancer cells. Hence, we carried out cell migration assay and knockdown experiments to clarify it (Figure 6A,6B). In si-SNHG15 knockdown MDA-MB-231 cells, the expression of miR-345-5p was significantly higher than that of miR-623 (Figure 6B), and thus, miR-345-5p was used for further analyses. Cell proliferation was significantly decreased after transfection with si-SNHG15, and this was restored by inhibiting the expression of miR-345-5p (Figure 6C). The similar result was observed with the cell migration assay (Figure 6A), where combined si-SNHG15 treatment and miR-345-5p inhibitor increased cell migration compared to si-SNHG15 alone (Figure 6D).
Discussion
Breast cancer is by far the most fatal malignant tumor globally, and the incidence continues to rise (29,30). Presently, clinical and pathologic characteristics are used to determine therapy and prognosis for breast cancer patients. Understanding the fundamental mechanisms that control breast cancer growth, and the development of new molecular biomarkers for the diagnosis and prognosis of breast cancer is critical. Despite new evidence that lnc-SNHG15 functions as an oncogene in a range of cancers (1,31), the specific mechanisms by which lnc-SNHG15 influences breast cancer development remain unclear. The current investigation demonstrated that the expression of lnc-SNHG15 is significantly enriched in breast cancer cells (MDA-MB-231) compared to normal breast epithelial cells (MCF10A). Based on the aforementioned results, we believe that lnc-SNHG15 is a promising and novel target for breast cancer therapy. Development of drug that can specifically target this lncRNA will benefit the prognosis of breast cancer.
Over the past decade, lncRNAs showed their promising potentials as the biomarkers as well as the therapeutic targets for multiple cancers (32). Small interfering RNAs and antisense oligonucleotides of lncRNAs were developed as biological agents and some of them were approved by FDA (33). Previous studies have demonstrated that SNHG15 influences numerous genes implicated in tumor formation, including CTGF, GADD45, MYC, and CASP3 (34-37). CTGF inhibits cell apoptosis and is responsible for chemoresistance to 5-fluorouracil (5-FU) (38). High levels of GADD45 expression make cells more resistant to UV irradiation or cisplatin, resulting in increased DNA repair and tumor development (35,39).
Our results also demonstrated that knocking down the expression of lnc-SNHG15 in breast cancer cells reduced cell proliferation, colony formation, and migration, whereas lnc-SNHG15 had the opposite effect. Chen et al. (40) found that suppressing lnc-SNHG15 following siRNA transfection inhibited cell proliferation, invasion, and triggered apoptosis via controlling the protein expression of metalloproteases (MMP2 and MMP9). MMPs are recognized to be involved in cell proliferation, differentiation, and angiogenesis, as well as cell metastasis (41). Our in vivo findings showed that silencing lnc-SNHG15 with si1-RNA reduced tumor development in the xenograft zebrafish model, and these findings were supported by previous publications (1,42).
Furthermore, we demonstrated that cisplatin treatment combined with SNHG15 silencing synergistically suppressed the proliferation and migration of breast cancer cells. Therefore, SNHG15 may be a key regulator in cisplatin resistance in breast cancer, and thus targeting SNHG15 may be an effective method for cisplatin chemoresistance in breast cancer. The potency of cisplatin has been linked to its capacity to crosslink with the purine bases on DNA to build DNA adducts, thereby blocking DNA repair and resulting in DNA damage, which ultimately leads to cancer cell death (43). However, there is still a paucity of studies examining the effects of silencing lnc-SNHG15 on cisplatin sensitivity in breast cancer cells. Besides, tumor-associated macrophages have been reported to be involved in cisplatin chemoresistance by changing the breast tumor microenvironment (44). According our in vivo results, SNHG15 silence increased the sensitivity of cisplatin in breast cancer zebrafish xenograft model, implying the critical roles of SNHG15 in cisplatin chemoresistance. However, whether tumor-associated macrophages regulate the cisplatin chemoresistance via SNHG15 in breast cancers requires further studies. Recent evidence suggested that SNHG15 could function as miRNA sponges to inhibit the expression of miRNAs (45). miRNAs play important roles in cell signaling, cell survival, DNA methylation, and invasiveness (46). MiR-345-5p has been shown to regulate the expression of human multidrug resistance-associated protein 1 (ABCC1/MRP1), suggesting that deregulation of miRNA expression is linked to the cisplatin resistant phenotype (46-48). In this study, we found that the effect of SNHG15 knockdown was restored by the inhibition of miR-345-5p, suggesting that overexpression miR-345-5p might play a key role in tumor suppression and cisplatin sensitivity. There is increasing evidence to suggest that miR-345-5p may act as a tumor suppressor in the tumorigenesis by regulating the expression of KISS1 and it may modulate metastasis by upregulating the production of E-cadherin (49,50). Moreover, downregulation of miR-345-5p has been implicated with chemoresistance in a range of malignancies (48,51).
Conclusions
The expression of lnc-SNHG15 was significantly increased in breast cancer cells, suggesting that lnc-SNHG15 may be a negative prognostic factor and a high risk factor for breast cancer patients. Silencing SNHG15 expression decreased the proliferation and migration ability of MDA-MB-231 cells. Moreover, cisplatin and lnc-SNHG15 knockdown acted synergistically to limit the progression of breast cancer. Lnc-SNHG15 knockdown may affect breast cancer progression via upregulating miR-345-5p expression. This research paves the way for additional research into lnc-SNHG15 and the miR-345-5p regulatory axis as potential therapeutic targets for breast cancer.
Acknowledgments
Funding: This study was funded by the Basic Public Welfare Research Project of Zhejiang Province (No. LGD20H090005); the Medical Science and Technology Project of Zhejiang Province (Nos. 2021KY1195, 2020KY348, 2021KY402); the Scientific Research Fund of Zhejiang Provincial Education Department (No. Y202146066); and the Taizhou Science and Technology Project (Nos. 20ywa11, 1902ky18).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5275/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-5275/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-5275/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All zebrafish experiments were approved by the Institutional Animal Care and Use Committee of Taizhou Hospital of Zhejiang Province Affiliated to Wenzhou Medical University (Linhai, China), in compliance with institutional guidelines for the care and use of animals. The project license No. EZY-2020038.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Du J, Zhong H, Ma B. Targeting a novel LncRNA SNHG15/miR-451/c-Myc signaling cascade is effective to hamper the pathogenesis of breast cancer (BC) in vitro and in vivo. Cancer Cell Int 2021;21:186. [Crossref] [PubMed]
- Jin L, Han B, Siegel E, et al. Breast cancer lung metastasis: Molecular biology and therapeutic implications. Cancer Biol Ther 2018;19:858-68. [Crossref] [PubMed]
- Wang R, Zhu Y, Liu X, et al. The Clinicopathological features and survival outcomes of patients with different metastatic sites in stage IV breast cancer. BMC Cancer 2019;19:1091. [Crossref] [PubMed]
- Jovanovic J, Rønneberg JA, Tost J, et al. The epigenetics of breast cancer. Mol Oncol 2010;4:242-54. [Crossref] [PubMed]
- Marino N, German R, Podicheti R, et al. Aberrant epigenetic and transcriptional events associated with breast cancer risk. Clin Epigenetics 2022;14:21. [Crossref] [PubMed]
- Liu L, Zhang Y, Lu J. The roles of long noncoding RNAs in breast cancer metastasis. Cell Death Dis 2020;11:749. [Crossref] [PubMed]
- Hu Y, Zhu QN, Deng JL, et al. Emerging role of long non-coding RNAs in cisplatin resistance. Onco Targets Ther 2018;11:3185-94. [Crossref] [PubMed]
- Zhong Y, Gao D, He S, et al. Dysregulated Expression of Long Noncoding RNAs in Ovarian Cancer. Int J Gynecol Cancer 2016;26:1564-70. [Crossref] [PubMed]
- Zhu Y, Zhao Y, Dong S, et al. Systematic identification of dysregulated lncRNAs associated with platinum-based chemotherapy response across 11 cancer types. Genomics 2020;112:1214-22. [Crossref] [PubMed]
- Zhang P, Wu W, Chen Q, et al. Non-Coding RNAs and their Integrated Networks. J Integr Bioinform 2019;16:20190027. [Crossref] [PubMed]
- Zhang X, Wang W, Zhu W, et al. Mechanisms and Functions of Long Non-Coding RNAs at Multiple Regulatory Levels. Int J Mol Sci 2019;20:5573. [Crossref] [PubMed]
- Shuai Y, Ma Z, Lu J, et al. LncRNA SNHG15: A new budding star in human cancers. Cell Prolif 2020;53:e12716. [Crossref] [PubMed]
- Zhang C, Ke Y, Liu X, et al. High Expression of the Long Noncoding RNA SNHG15 in Cancer Tissue Samples Predicts an Unfavorable Prognosis of Cancer Patients: A Meta-Analysis. J Oncol 2020;2020:3417036. [Crossref] [PubMed]
- Zimta AA, Tigu AB, Braicu C, et al. An Emerging Class of Long Non-coding RNA With Oncogenic Role Arises From the snoRNA Host Genes. Front Oncol 2020;10:389. [Crossref] [PubMed]
- Liu K, Hou Y, Liu Y, et al. LncRNA SNHG15 contributes to proliferation, invasion and autophagy in osteosarcoma cells by sponging miR-141. J Biomed Sci 2017;24:46. [Crossref] [PubMed]
- O'Brien J, Hayder H, Zayed Y, et al. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne) 2018;9:402. [Crossref] [PubMed]
- Wahid F, Shehzad A, Khan T, et al. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta 2010;1803:1231-43. [Crossref] [PubMed]
- Otmani K, Lewalle P. Tumor Suppressor miRNA in Cancer Cells and the Tumor Microenvironment: Mechanism of Deregulation and Clinical Implications. Front Oncol 2021;11:708765. [Crossref] [PubMed]
- Zhang J, Wang C, Yan S, et al. miR-345 inhibits migration and stem-like cell phenotype in gastric cancer via inactivation of Rac1 by targeting EPS8. Acta Biochim Biophys Sin (Shanghai) 2020;52:259-67. [Crossref] [PubMed]
- Chen L, Li X, Chen X. Prognostic significance of tissue miR-345 downregulation in non-small cell lung cancer. Int J Clin Exp Med 2015;8:20971-6. [PubMed]
- Yu H, Li SB. Role of LINC00152 in non-small cell lung cancer. J Zhejiang Univ Sci B 2020;21:179-91. [Crossref] [PubMed]
- Hosono Y, Niknafs YS, Prensner JR, et al. Oncogenic Role of THOR, a Conserved Cancer/Testis Long Non-coding RNA. Cell 2017;171:1559-1572.e20. [Crossref] [PubMed]
- Wang J, Zhang XY, Xu DY. Zebrafish xenograft model for studying the function of lncRNA SNHG4 in the proliferation and migration of colorectal cancer. J Gastrointest Oncol 2022;13:210-20. [Crossref] [PubMed]
- Weinberg MS, Morris KV. Long non-coding RNA targeting and transcriptional de-repression. Nucleic Acid Ther 2013;23:9-14. [Crossref] [PubMed]
- Aman A, Piotrowski T. Cell migration during morphogenesis. Dev Biol 2010;341:20-33. [Crossref] [PubMed]
- Marques IJ, Weiss FU, Vlecken DH, et al. Metastatic behaviour of primary human tumours in a zebrafish xenotransplantation model. BMC Cancer 2009;9:128. [Crossref] [PubMed]
- Ni W, Yao S, Zhou Y, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol Cancer 2019;18:143. [Crossref] [PubMed]
- Wang L, Zhao X, Fu J, et al. The Role of Tumour Metabolism in Cisplatin Resistance. Front Mol Biosci 2021;8:691795. [Crossref] [PubMed]
- Sharma GN, Dave R, Sanadya J, et al. Various types and management of breast cancer: an overview. J Adv Pharm Technol Res 2010;1:109-26. [PubMed]
- Nagai H, Kim YH. Cancer prevention from the perspective of global cancer burden patterns. J Thorac Dis 2017;9:448-51. [Crossref] [PubMed]
- Xu B, Mei J, Ji W, et al. LncRNA SNHG3, a potential oncogene in human cancers. Cancer Cell Int 2020;20:536. [Crossref] [PubMed]
- Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res 2017;77:3965-81. [Crossref] [PubMed]
- Winkle M, El-Daly SM, Fabbri M, et al. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov 2021;20:629-51. [Crossref] [PubMed]
- Yin D, Chen W, O'Kelly J, et al. Connective tissue growth factor associated with oncogenic activities and drug resistance in glioblastoma multiforme. Int J Cancer 2010;127:2257-67. [Crossref] [PubMed]
- Saeinasab M, Bahrami AR, González J, et al. SNHG15 is a bifunctional MYC-regulated noncoding locus encoding a lncRNA that promotes cell proliferation, invasion and drug resistance in colorectal cancer by interacting with AIF. J Exp Clin Cancer Res 2019;38:172. [Crossref] [PubMed]
- Salvador JM, Brown-Clay JD, Fornace AJ Jr. Gadd45 in stress signaling, cell cycle control, and apoptosis. Adv Exp Med Biol 2013;793:1-19. [Crossref] [PubMed]
- Devarajan E, Sahin AA, Chen JS, et al. Down-regulation of caspase 3 in breast cancer: a possible mechanism for chemoresistance. Oncogene 2002;21:8843-51. [Crossref] [PubMed]
- Yang K, Gao K, Hu G, et al. CTGF enhances resistance to 5-FU-mediating cell apoptosis through FAK/MEK/ERK signal pathway in colorectal cancer. Onco Targets Ther 2016;9:7285-95. [Crossref] [PubMed]
- Smith ML, Kontny HU, Zhan Q, et al. Antisense GADD45 expression results in decreased DNA repair and sensitizes cells to u.v.-irradiation or cisplatin. Oncogene 1996;13:2255-63. [PubMed]
- Chen SX, Yin JF, Lin BC, et al. Upregulated expression of long noncoding RNA SNHG15 promotes cell proliferation and invasion through regulates MMP2/MMP9 in patients with GC. Tumour Biol 2016;37:6801-12. [Crossref] [PubMed]
- Quintero-Fabián S, Arreola R, Becerril-Villanueva E, et al. Role of Matrix Metalloproteinases in Angiogenesis and Cancer. Front Oncol 2019;9:1370. [Crossref] [PubMed]
- Kong Q, Qiu M. Long noncoding RNA SNHG15 promotes human breast cancer proliferation, migration and invasion by sponging miR-211-3p. Biochem Biophys Res Commun 2018;495:1594-600. [Crossref] [PubMed]
- Dasari S, Tchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol 2014;740:364-78. [Crossref] [PubMed]
- Li H, Yang P, Wang J, et al. HLF regulates ferroptosis, development and chemoresistance of triple-negative breast cancer by activating tumor cell-macrophage crosstalk. J Hematol Oncol 2022;15:2. [Crossref] [PubMed]
- Mi H, Wang X, Wang F, et al. SNHG15 Contributes To Cisplatin Resistance In Breast Cancer Through Sponging miR-381. Onco Targets Ther 2020;13:657-66. [Crossref] [PubMed]
- Natesh NS, White BM, Bennett MMC, et al. Emerging Role of miR-345 and Its Effective Delivery as a Potential Therapeutic Candidate in Pancreatic Cancer and Other Cancers. Pharmaceutics 2021;13:1987. [Crossref] [PubMed]
- Pogribny IP, Filkowski JN, Tryndyak VP, et al. Alterations of microRNAs and their targets are associated with acquired resistance of MCF-7 breast cancer cells to cisplatin. Int J Cancer 2010;127:1785-94. [Crossref] [PubMed]
- Hu W, Tan C, He Y, et al. Functional miRNAs in breast cancer drug resistance. Onco Targets Ther 2018;11:1529-41. [Crossref] [PubMed]
- Ulasov I, Borovjagin A, Fares J, et al. MicroRNA 345 (miR345) regulates KISS1-E-cadherin functional interaction in breast cancer brain metastases. Cancer Lett 2020;481:24-31. [Crossref] [PubMed]
- Na TY, Schecterson L, Mendonsa AM, et al. The functional activity of E-cadherin controls tumor cell metastasis at multiple steps. Proc Natl Acad Sci U S A 2020;117:5931-7. [Crossref] [PubMed]
- Mou T, Xie F, Zhong P, et al. MiR-345-5p functions as a tumor suppressor in pancreatic cancer by directly targeting CCL8. Biomed Pharmacother 2019;111:891-900. [Crossref] [PubMed]
(English Language Editor: J. Teoh)


