
Exploration of the shared gene signatures and molecular mechanisms between atherosclerosis and rheumatoid arthritis via multi-microarray data analyses
Introduction
Atherosclerosis (AS) is recognized as a slow, aggressive non-bacterial inflammatory disorder within the arterial wall, involving endothelial dysfunction, vascular smooth muscle cells, macrophage migration, phenotypic transformation, lipid deposit, plaque rupture, and consequential acute ischemic vascular events. Rheumatoid arthritis (RA) is a chronic systemic autoimmune inflammatory disease which contributes an augmented risk of cardiovascular diseases (CVD), including myocardial infarction and stroke (1), and represents a tremendous socioeconomic burden worldwide. Accumulating evidence indicates the RA inflammatory state predisposes to the acceleration of AS (2,3). Not only are shared their risk factors between RA and AS, such as smoking, diabetes mellitus, obesity, and genetic risk factors, the two disorders share many overlapping immune environmental factors, including inflammatory cytokines/chemokines, and local and systemic immune reactions involved signaling pathways. Multi-aspects of the pathophysiological process of AS can be mirrored in diseased RA synovium, including distinct immune cell infiltration, extracellular matrix remodeling, and neovascularization. Nevertheless, the potential mechanisms of accelerating AS in RA have not been fully elucidated.
The small sample size and lack of clinical significance of previous studies produced the present study. In this study, a comprehensive bioinformatics analysis was implemented to probe the underlying relevance of the pathogenesis of RA complicated with AS. Firstly, we integrated transcriptional profiling of synovial tissues from both RA and atherosclerotic plaques from AS, and identified their common differentially expressed genes (DEGs) and hub genes. Secondly, we probed their function and involved molecular mechanisms to investigate whether and how RA plays a causative role in AS progression. Finally, exploitation of candidate drugs targeting hub genes was conducted, which may hold great promise in the future fight against the comorbidities. To our knowledge, few comprehensive analyses of RA and AS via multi-microarray data analyses have been reported. We present the following article in accordance with the STREGA reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4934/rc).
Methods
Datasets collection
The GSE55235 and GSE55457 datasets expound the transcriptional profiling of synovial tissues in patients with RA compared with healthy individuals, and the GSE28829 and GSE41571 datasets elucidate the specifical transcriptional signatures of advanced atherosclerotic plaques compared with early AS. The mRNA expression of hub genes subsequently identified were verified in the GSE77298 and GSE163154 datasets, respectively. The flowchart for the current research is shown in Figure 1. All datasets were extracted from the GEO database of the National Center for Biotechnology Information platform (https://www.ncbi.nlm.nih.gov/geo) (4,5), and information on the platform, sample, source tissue, contributors, and attributions for the six datasets is listed in Table 1. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
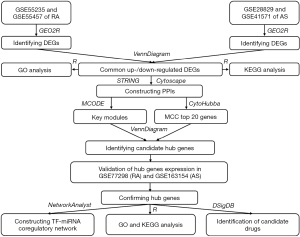
Table 1
| GEO accession | Platform | Samples [number] | Source tissue | Contributors | Attribute |
|---|---|---|---|---|---|
| GSE55235 | GPL96 | Rheumatoid arthritis [10]; healthy control [10] | Human synovium | Woetzel D, Huber R, Kupfer P, et al. | Test set |
| GSE55457 | GPL96 | Rheumatoid arthritis [13]; healthy control [10] | Human synovium | Woetzel D, Huber R, Kupfer P, et al. | Test set |
| GSE28829 | GPL570 | Advanced atherosclerotic plaque [16]; early atherosclerotic plaque [13] | Human carotid atheroma | Manca M, Biessen E, Daemen M | Test set |
| GSE41571 | GPL570 | Unstable atheromatous lesions [ruptured, 5]; stable atheromatous lesions [unruptured, 6] | Human carotid atheroma | Lee K, Santibanez-Koref M, Polvikoski T, et al. | Test set |
| GSE77298 | GPL570 | Rheumatoid arthritis [16]; healthy control [7] | Human synovium | Broeren MG | Validation set |
| GSE163154 | GPL6104 | High-risk atherosclerotic lesion [intraplaque haemorrhage, 27]; low-risk atherosclerotic lesion [non-intraplaque haemorrhage, 16] | Human carotid atheroma | Jin H, Biessen E | Validation set |
GPL96, (HG-U133A) Affymetrix Human Genome U133A Array; GPL570, (HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array; GPL6104, Illumina humanRef-8 v2.0 expression beadchip.
Identification of DEGs and common DEGs between RA and AS
The original files (cel format) downloaded from the GEO database were pre-processed and normalized by the Robust Multiarray Average (RMA) method based on the R software (version 3.6.3) Affy package (6). Probe sets with no corresponding gene symbol or genes with more than one probe set were removed or averaged, respectively, and the ComBat function of the SVA package in R was used to remove the batch effect. After background correction, quality control, standardization, and ID transformation from probes to gene symbols, we used GEO2R to identify the DEGs between the different groups by comparing their gene expression profile. GEO2R (7) (https://www.ncbi.nlm.nih.gov/geo/ge2r) is a web-accessible analysis tool based on two R packages, namely GEOquery (8) and limma (9), the former to read data, and the latter to calculate the differential expression multiple. The cut-off criteria for statistical significance to identify DEGs was set as adjusted P value <0.05 and |logfold change (FC)|≥1. The DEGs were fully visualized in a volcano plot and partially in a heat map using the R package ‘ggplot2’ (https://cran.r-project.org/web/packages/ggplot2/) and ‘complexHeatmap’ (10), respectively. The identification of common DEGs between RA and AS was acquired by the R programming language and visualized through the ‘VennDiagram’ (https://cran.r-project.org/web/packages/VennDiagram/).
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses
GO (11,12) and KEGG (13,14) pathway enrichment analyses of common DEGs or hub genes were performed by the R package ‘clusterProfiler’ (http://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html) (15). The enrichment analysis results of GO categories included biological process (BP), cellular component (CC), and molecular function (MF) (16). To illustrate and understand the metabolic or signaling pathways which the DEGs might be involved in, KEGG pathway enrichment analysis was simultaneously implemented, and the significance threshold was adjusted P value <0.05.
Protein-protein interactions (PPI) network and key module construction
PPI networks can provide abundant information on the functional interactions between proteins in cellular biology studies (17,18). The common DEGs were input into Search Tool for the Retrieval of Interacting Genes (STRING) (https://string-db.org/) online for creating a PPI network (19). STRING transmits experimental and predictive interaction-founded information and the interaction defined with 3-dimensional structures, auxiliary biomedical information, and confidence score (20). The confidence score in the current study was set as 0.400, a middle value utilizing the STRING platform. Subsequently, the acquired PPIs network was reimported into Cytoscape for further analysis (https://cytoscape.org/) (21,22), and Cytoscape’s plug-in molecular complex detection technology (MCODE) was applied to probe key functional modules. The selection criteria weres set as: K-core =2, degree cutoff =2, max depth =100, and node score cutoff =0.2.
Candidate hub gene identification
The Cytoscape software plugin cytoHubba uses multiple scoring methods to dissect the PPIs network. The top 20 genes chosen by the topological algorithm of maximum correlation criterion (MCC) were overlapped with genes in the key modules, and the overlapped genes were identified as candidate hub genes.
Validation of hub gene expression in external datasets and enrichment analyses
The mRNA expression level of candidate hub genes was verified in the GSE77298 and GSE163154 datasets, respectively. The GSE77298 dataset included 16 RA and seven healthy controls, while the GSE163154 dataset contained 27 high-risk atherosclerotic lesion (intraplaque haemorrhage) and 16 low-risk atherosclerotic lesion (non-intraplaque haemorrhage), which indicated the different stages of AS progression. Student’s t-test was used for group comparisons of continuous variables distributed normally, while the Mann-Whitney U-test was used for variables with an abnormal distribution. The significance threshold was P value <0.05. The aforementioned candidate hub genes that were simultaneously differentially expressed in the GSE77298 and GSE163154 datasets were confirmed as hub genes. Thereafter the GO and KEGG pathway analyses of confirmed hub genes were executed via R package ‘clusterProfiler’.
Transcription factor (TF)-miRNA coregulatory network
The reciprocity of TFs and miRNAs with the hub genes may play a vital role in regulating the expression of hub genes. The TF-miRNA coregulatory network for hub genes was established using NetworkAnalyst (https://www.networkanalyst.ca/), a powerful online-based platform for comprehensive analyses, systematic interpretation, and deciphering of gene expression (23,24) and RegNetwork repository, a synthetical database of transcriptional and post-transcriptional regulating networks (25).
Identification of candidate drugs
The identification of candidate drugs for treating RA combined with AS focused on the aforementioned confirmed hub genes. The user-friendly web-based enrichment analysis platform Enrichr (26) furnishes free access to the Drug Signatures database (DSigDB, http://biotechlab.fudan.edu.cn/database/drugsig/). DSigDB (27) is an absolute drug response gene signature database assembling drug response microarray data and annotated drug and target information derived from public databases and scientific documentation and was established by singling out the top 500 upregulated and downregulated genes as drug signatures. Current DrugSig includes more than 1,300 drugs, 7,000 microarrays, and 800 targets, enabling computational drug repositioning to exploit neoteric drugs that may target hub genes. The cut-off criterion was P-adjusted <0.05.
Results
Identification of DEGs
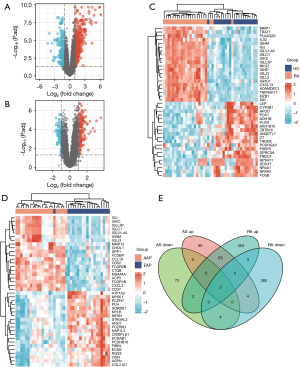
Following standardization there was a fundamentally same average gene expression value for each sample, and the differences between groups were examined with principal component analysis (PCA), indicating the sample data were credible. The data pre-processing is presented in Figure S1. After comprehensive analysis of RA-related datasets GSE55235 and GSE55457, 418 DEGs were upregulated and 268 were downregulated, while for the AS-related datasets GSE28829 and GSE41571, 162 DEGs were upregulated and 79 downregulated. The expression profiling for the two diseases is visualized in a volcano plot (Figure 2A,2B), respectively, and that of the top 20 genes with high and low expression level is visualized in a heat map (Figure 2C,2D), respectively. As shown in Figure 2E, following overlap of these DEGs there were 63 common upregulated and four common downregulated DEGs between RA and AS.
GO and KEGG pathway enrichment analyses of common DEGs
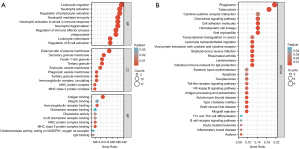
GO and KEGG pathway enrichment analyses were conducted to probe the biological functions and pathways involved in RA and AS of the 67 common DEGs, and the top 10 terms of each category (BP, CC and MF) are visualized in the bubble diagram (Figure 3A). KEGG analysis is shown in Figure 3B, and GO and KEGG analyses information are presented in Table S1 in detail. The BP subset indicated common DEGs were highly enhanced in leukocyte migration, regulation of lymphocyte activation, neutrophil activation, and phagocytosis, and the CC subsection mainly manifested external side of plasma membrane, tertiary granule, secretory granule membrane, and endocytic vesicle membrane. The MF part expounded antigen binding, MHC class II protein complex, and immunoglobulin receptor binding and chemokine activity. KEGG pathway enrichment analyses showed common DEGs were significantly involved in phagosome, viral myocarditis, tuberculosis, intestinal immune network for IgA production, hematopoietic cell lineage, cell adhesion molecules, rheumatoid arthritis, leukocyte transendothelial migration, the chemokine signaling pathway, the NF-κB signaling pathway, the Toll-like receptor signaling pathway, apoptosis, and Th1 and Th2 cell differentiation.
PPI network and key module construction
A PPI network was established for further analysis including module analysis, hub genes identification, and TF-miRNA coregulatory network construction, and the network contained 52 nodes and 229 edges. To probe key functional modules, MCODE was applied, and two modules were generated (Figure 4A). Detail information is listed in Table S2.

Candidate hub gene identification
CytoHubba was applied, and the top 20 genes based on MCC scoring are shown and highlighted in a sub-network with 46 nodes and 220 edges (Figure 4B). To obtain candidate hub genes, the 20 genes were overlapped with those in the key modules. The overlapped genes were identified as candidate hub genes, namely PTPRC, ITGB2, CD86, CSF1R, CTSS, TLR8, CD14, MMP9, S100A9, CD48, FCGR2B, CSF2RB, LCP1, DOCK2, RAC2, EVI2B, CD52, HLA-DRA, CD300A, and NCKAP1L, and the details are presented in Table 2.
Table 2
| No. | Gene symbol | Full name of protein | Main function |
|---|---|---|---|
| 1 | PTPRC | Receptor-type tyrosine-protein phosphatase C | Protein tyrosine-protein phosphatase required for T-cell activation through the antigen receptor; acts as a positive regulator of T-cell coactivation upon binding to DPP4 |
| 2 | ITGB2 | Integrin beta-2 | Integrin ITGAL/ITGB2 is a receptor for ICAMs and the secreted form of ubiquitin-like protein ISG15; the interaction is mediated by ITGA; receptors for the iC3b fragment of the third complement component and for fibrinogen; integrin ITGAX/ITGB2 recognizes the sequence G-P-R in fibrinogen alpha-chain; integrin ITGAD/ITGB2 is a receptor for ICAM3 and VCAM1, contributes to natural killer cell cytotoxicity; involved in leukocyte adhesion and transmigration of leukocytes; integrin ITGAL/ITGB2 in association with ICAM3 contributes to apoptotic neutrophil phagocytosis by macrophages |
| 3 | CD86 | T-lymphocyte activation antigen CD86 | Receptor involved in the costimulatory signal essential for T-lymphocyte proliferation and interleukin-2 production; plays a critical role in the early events of T-cell activation and costimulation of naive T-cells; involved in the regulation of B cells function and plays a role in regulating the level of IgG1 produced; upon CD40 engagement, activates NF-kappa-B signaling pathway via phospholipase C and protein kinase C activation |
| 4 | CSF1R | Macrophage colony-stimulating factor 1 receptor | Tyrosine-protein kinase that acts as cell-surface receptor for CSF1 and IL34 and plays an essential role in the regulation of survival, proliferation, and differentiation of hematopoietic precursor cells, especially mononuclear phagocytes, such as macrophages and monocytes; promotes the release of proinflammatory chemokines in response to IL34 and CSF1 in innate immunity; promotes reorganization of the actin cytoskeleton, regulates formation of membrane ruffles, cell adhesion, and cell migration, and promotes cancer cell invasion; activates several signaling pathways in response to ligand binding, including the ERK1/2 and the JNK pathway |
| 5 | CTSS | Cathepsin S | Thiol protease, key protease responsible for the removal of the invariant chain from MHC class II molecules and MHC class II antigen presentation; the bond-specificity of this proteinase is in part similar to the specificities of cathepsin L |
| 6 | TLR8 | Toll-like receptor 8 | Endosomal receptor that plays a key role in innate and adaptive immunity; controls host immune response against pathogens through recognition of RNA degradation products specific to microorganisms; recognizes GU-rich single-stranded RNA derived from SARS-CoV-2, SARS-CoV-1, and HIV-1 viruses; upon binding to agonists, undergoes dimerization that brings TIR domains from the two molecules into direct contact, leading to the recruitment of TIR-containing downstream adapter MYD88 through homotypic interaction; in turn, the Myddosome signaling complex is formed involving IRAK4, IRAK1, TRAF6, and TRAF3 leading to activation of downstream transcription factors NF-kappa-B and IRF7 to induce proinflammatory cytokines and interferons, respectively |
| 7 | CD14 | Monocyte differentiation antigen CD14 | Coreceptor for bacterial lipopolysaccharide; mediates the innate immune response to bacterial lipopolysaccharide; acts via MyD88, TIRAP, and TRAF6, leading to NF-kappa-B activation, cytokine secretion, and the inflammatory response; binds electronegative LDL and mediates the cytokine release |
| 8 | MMP9 | Matrix metalloproteinase-9 | Matrix metalloproteinase that plays an essential role in local proteolysis of the extracellular matrix and in leukocyte migration; cleaves type IV and type V collagen into large C-terminal three quarter fragments and shorter N-terminal one quarter fragments; degrades fibronectin |
| 9 | S100A9 | Protein S100-A9 | Regulates neutrophil number and apoptosis by an anti-apoptotic effect; regulates cell survival via ITGAM/ITGB and TLR4 and a signaling mechanism involving MEK-ERK |
| 10 | CD48 | CD48 antigen | Ligand for CD2; might facilitate interaction between activated lymphocytes; probably involved in regulating T-cell activation |
| 11 | FCGR2B | Low affinity immunoglobulin gamma Fc region receptor II-b | Receptor for the Fc region of complexed or aggregated gamma immunoglobulins; involved in a variety of effector and regulatory functions such as phagocytosis of immune complexes and modulation of antibody production by B-cells; binds to this receptor results in down-modulation of previous state of cell activation triggered via antigen receptors on B-cells (BCR), T-cells (TCR), or via another Fc receptor |
| 12 | CSF2RB | Cytokine receptor common subunit beta | High affinity receptor for interleukin-3, interleukin-5, and granulocyte-macrophage colony-stimulating factor |
| 13 | LCP1 | Plastin-2 | Actin-binding protein; plays a role in the activation of T-cells in response to costimulation through TCR/CD3 and CD2 or CD28; modulates the cell surface expression of IL2RA/CD25 and CD69 |
| 14 | DOCK2 | Dedicator of cytokinesis protein 2 | Involved in cytoskeletal rearrangements required for lymphocyte migration in response to chemokines; activates RAC1 and RAC2 by functioning as a guanine nucleotide exchange factor; participates in IL2 transcriptional activation via the activation of RAC2 |
| 15 | RAC2 | Ras-related C3 botulinum toxin substrate 2 | Plasma membrane-associated small GTPase which cycles between an active GTP-bound and inactive GDP-bound state; in the active state binds to a variety of effector proteins to regulate cellular responses, such as secretory processes, phagocytose of apoptotic cells, and epithelial cell polarization; augments the production of ROS by NADPH oxidase |
| 16 | EVI2B | Protein EVI2B | Required for granulocyte differentiation and functionality of hematopoietic progenitor cells through the control of cell cycle progression and survival of hematopoietic progenitor cells |
| 17 | CD52 | CAMPATH-1 antigen | May play a role in carrying and orienting carbohydrate, as well as having a more specific role. |
| 18 | HLA-DRA | HLA class II histocompatibility antigen, DR alpha chain | An alpha chain of antigen-presenting MHC II molecule; in complex with the beta chain HLA-DRB, displays antigenic peptides on professional APCs for recognition by TCR on HLA-DR-restricted CD4-positive T cells; guides antigen-specific T-helper effector functions; presents peptides derived from intracellular proteins that are trapped in autolysosomes after macroautophagy |
| 19 | CD300A | CMRF35-like molecule 8 | Inhibitory receptor which may contribute to the downregulation of cytolytic activity in natural killer cells, and to the downregulation of mast cell degranulation; negatively regulates the TLR signaling mediated by MYD88 but not TRIF through activation of PTPN6 |
| 20 | NCKAP1L | Nck-associated protein 1-like | Essential hematopoietic-specific regulator of the actin cytoskeleton; controls lymphocyte development, activation, proliferation, and homeostasis, erythrocyte membrane stability, as well as phagocytosis and migration by neutrophils and macrophages; required for efficient T-lymphocyte and neutrophil migration; involved in mechanisms WAVE-independent to regulate myosin and actin polymerization during neutrophil chemotaxis |
LDL, low-density lipoprotein; GTP, guanosine triphosphate; GDP, guanosine diphosphate; ROS, reactive oxygen species; NADPH, nicotinamide adenine dinucleotide phosphate; APCs, antigen-presenting cells.
Validation of hub gene expression in external datasets and enrichment analyses
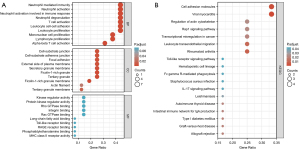
The mRNA expression level of 20 hub genes were verified in the RA-related dataset GSE77298 and AS-related dataset GSE163154, respectively. Compared with normal synovial tissues, most hub genes were significantly upregulated in the synovium of RA except for four genes, namely CTSS, CD14, CD48, and FCGR2B (Figure 5). Although the four genes had an ascending tendency in RA, they were excluded from hub genes. Similarly, the expression of all hub genes in high-risk atherosclerotic lesions with intraplaque haemorrhage was also higher than in low-risk atherosclerotic lesions without intraplaque haemorrhage (Figure 6). Accordingly, 16 hub genes were confirmed and subject to further analysis. GO enrichment analyses illustrated hub genes were significantly involved in leukocyte proliferation, leukocyte cell-cell adhesion, T cell activation, secretory granule membrane, external side of plasma membrane, focal adhesion, Rac GTPase binding, integrin binding, and MHC class II receptor activity, while KEGG pathway enrichment analysis revealed hub gens were significantly involved in viral myocarditis, cell adhesion molecules, rheumatoid arthritis, leukocyte transendothelial migration, the Rap1 signaling pathway, regulation of actin cytoskeleton, IL-17 signaling pathway, Fc gamma R-mediated phagocytosis, and the Toll-like receptor signaling pathway. GO and KEGG analyses outcomes are partially visualized in a bubble diagram (Figure 7A,7B), and Table S3 displays the analyses in detail.


TF-miRNA coregulatory network
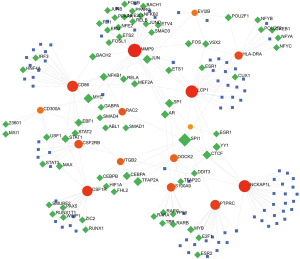
The TF-miRNA coregulatory network is shown in Figure 8, constituting 176 nodes (15 hub genes included) and 214 edges and includes abundant interactions between TFs, miRNAs, and hub genes. TFs, namely SPI1, MYC, JUN, TFAP2A, SP1, and CTCF interacted with 11, 5, 4, 4, 4, and 4 hub genes, respectively. Hsa-miR-204, hsa-miR-377, hsa-miR-493, and hsa-miR-525-5p respectively interacted with two hub genes simultaneously.
Identification of candidate drugs
Candidate drugs are depicted in Table 3, namely simvastatin, 5-azacytidine, bisindolylmaleimide, retinoic acid, and verteporfin.
Table 3
| Term | P value | Adjusted P value | Genes |
|---|---|---|---|
| Simvastatin CTD 00007319 | 8.27E-08 | 7.00E-05 | CD86; ITGB2; RAC2; HLA-DRA; MMP9; S100A9 |
| 5-azacytidine CTD 00005455 | 6.11E-06 | 0.00103537 | PTPRC; ITGB2; HLA-DRA; MMP9; S100A9 |
| Bisindolylmaleimide i CTD 00002749 | 1.62E-05 | 0.001501768 | CD86; ITGB2; MMP9 |
| Retinoic acid CTD 00006918 | 5.99E-05 | 0.003227657 | CD86; CSF1R; CD52; PTPRC; CD300A; ITGB2; RAC2; HLA-DRA; EVI2B; MMP9; S100A9 |
| Verteporfin HL60 DOWN | 6.25E-05 | 0.003227657 | PTPRC; CD300A; ITGB2; CSF2RB |
Discussion
A growing body of research has demonstrated inflammation builds a bridge between RA and AS. However, the underlying mechanisms of the pathogenesis of AS in RA patients remains unclear. In the present study, we analyzed several microarray data by a series of bioinformatics methods and attempted to comprehensively demonstrate the pathogenesis involved and provide new therapeutic targets for AS in RA.
Firstly, we identified 67 common DEGs between RA and AS and explored biological functions of these genes through GO and KEGG pathway enrichment analyses. The GO analysis results were concentrated at inflammatory cells-mediated inflammatory immune responses, which indicated inflammation played an essential role in the initial period of AS in RA. RA is recognized as a chronic autoimmune disease characterized by progressive systemic inflammation. Various immune cells are involved in the pathomechanism of RA, such as DCs, T cells, B cells, and macrophages, through releasing pro-inflammatory cytokines, secreting autoantibodies, and triggering the complement cascade, which promotes the establishment and maintenance of an inflammatory state (28). Coincidentally, these immune cells and inflammatory processes also take part in the pathogenesis of AS (29,30). This evidence lends support to the hypothesis that auto immune and inflammation result in the initiation and progression of atherosclerotic plaque in patients with RA. KEGG analysis also drew our attention to cytokine-cytokine receptor interactions. Many kinds of cytokines have been proven to participate in the establishment of inflammation in RA, such as tumor necrosis factor-α, interleukin-17A, and interferon-γ (IFN-γ) (31). These cytokines can cause cartilage and bone pathological damage through enhancing antigen presentation and macrophage activation, inducing inflammatory cells differentiation and recruitment, other inflammatory cytokines, and chemokine production (28). These cytokines and associated signaling pathways are also part of the pathogenesis of AS (32) and include IFN-γ, which aggravated neointimal hyperplasia and cholesterol accumulation by inducing endoplasmic reticulum stress and apoptosis, which accelerated AS (33,34). Cytokine-cytokine receptor interaction probably takes part in the development of AS in RA patients. In addition, cell adhesion molecules are indicated as an important pathway in the relationship between RA and AS. Adhesion molecules have been shown be highly expressed in RA synovial tissue cells, especially junctional adhesion molecule (JAM), intracellular adhesion molecule (ICAM), and vascular cell adhesion molecular (VCAM) (35). Recent studies have reported JAM-C was increased in an AS mouse model, and decreasing its expression could attenuate atherogenesis through reducing neointimal hyperplasia and monocyte recruitment (36-38). These results indicate JAM-C might provide a novel molecular target in AS, and treatment with anti-JAM-C antibody might ameliorate AS in RA patients. In addition, the Toll-like receptor signaling pathway has been reported to be a potential link between RA and AS (39), and our results also found it increased risk the risk of AS in RA patients. These pathways might account for the relationship between RA and AS and could be studied subsequently.
Secondly, to investigate the pathogenesis of AS in RA, we identified 20 hub genes based on common DEGs through PPI network and key module construction. Moreover, we successfully verified 16 hub genes in other datasets to improve the credibility and universality of our results. The reason why the four hub genes were not validated may be because of the relatively small sample size of the external RA dataset. CD86 is a co-stimulatory molecule in the cell membrane of dendritic cells (DCs) and plays an important immunomodulatory function in T cell activation (40,41). It has been reported that the differentiation and maturation of DCs with the upregulation of CD86 might accelerate RA, and importantly, researchers have found low-density lipoprotein (LDL) and oxidized LDL (oxLDL), which trigger the development of AS, could induce the expression of CD86 (42). On the contrary, reducing the level of CD86 and subsequent DCs maturation could be a therapeutic target for both RA and AS (43). In our current study, CD86 was upregulated in these diseases, which is consistent with previous studies (42,43) and indicated it was probably a key molecule linking the two. MMP9 is considered an enhancer of extracellular matrix destruction, and its level is higher in unstable plaques. It has been reported that MMP9 levels were up-regulated in RA, which partly explains the AS burden and plaque vulnerability in the disease (44-46). Nevertheless, drugs that reduce its expression level have the therapeutic potential to prevent AS in RA patients (47,48). In addition, TLR8 and S100A9 have been indicated as mediators between RA and AS (49,50), and other hub genes related with either have been reported, although further research is required to explore their role in the pathogenesis. These genes might be involved in the development of AS in RA and could become preventive and therapeutic targets. Similarly, we performed GO and KEGG pathway enrichment analyses of the 16 hub genes mentioned previously, and the results were the same as those found with common DEGs. In addition to the previously mentioned pathways, the Rap1 signaling pathway was one of the top items in enrichment analyses. Recent studies have indicated suppression of Rap1 might contribute to the pathogenesis of RA (51-53), and the Rap1 signaling pathway offers protection against AS progression through stimulating nitric oxide (NO) release and restricting proinflammatory signaling (54). However, a previous study has found the activity of Rap1 was up-regulated in AS (55), and the role of the Rap1 signaling pathway in RA-induced AS requires further study.
Targeting the regulatory elements of common and hub genes is also a therapeutic option. Therefore, we constructed a TF-miRNA coregulatory network to investigate upstream molecules of hub genes. MYC is an important transcription factor that tightly correlates to cell proliferation (56), and many researchers have found its level is up-regulated in RA and that drug treatment could inhibit its expression (57,58). Consistent with this trend, the expression of MYC was increased in AS lesions, and decreasing its level could reduce the pathogenesis of AS (59,60). These results indicated targeted therapy to MYC might prevent AS in RA patients. Although other transcription factors or miRNAs have not been related to both RA and AS yet, our study could provide a direction for further research. For instance, hsa-miR-204 was downregulated in RA as a target of long non-coding RNA NEAT1, and decreasing its expression level was related to the promotion of cell proliferation, inflammatory cytokine production, and the attenuation of cell apoptosis (61,62). Several pathological processes of AS have been reported as being influenced by hsa-miR-204, including vascular and valves calcification, ageing, foam cell formation, inflammation, apoptosis, endothelial cell dysfunction, and vascular smooth muscle cell proliferation and migration (63-67). These and other results suggest hsa-miR-204 probably acts as a key molecule in the development of AS in RA and deserves further study. In addition, hsa-miR-377, hsa-miR-493, and hsa-miR-525-5p have been reported in either RA or AS (68-73), and whether these miRNAs take part in the other disease is also worthy of further investigation.
Lastly, we identified candidate drugs based on hub genes by bioinformatic methods to treat patients with RA and AS. Apart from classical anti-atherosclerotic drugs, such as simvastatin, several newfound therapeutic options attracted our attention, including 5-azacytidine, a DNA methyltransferase-1 inhibitor, which could exert a protective effect from AS through promoting the maintenance of vascular smooth muscle cells, reducing pathological vascular remodeling, and decreasing inflammatory cytokine expression (74). Another study also reported 5-azacytidine reduced disturbed flow-induced endothelial inflammation and the development of AS in a mouse model (75), and this beneficial effect could also be achieved by suppressing macrophage inflammation (76). DNA hypermethylation also plays an essential role in the pathogenesis of RA, and 5-azacytidine could ameliorate its progression (77,78). Therefore, treatment of RA with 5-azacytidine might not only improve RA but also slow the development of AS in RA patients. Retinoic acid exhibited its beneficial effect on RA through inhibiting the inflammatory response, suppressing cell motility, migration, and invasiveness, and inducing apoptosis (79,80), and the retinoic acid signaling pathway is dysregulated in AS (81,82). To sum up, these candidate drugs could be therapeutic for patients with RA and AS.
Recently, several researches attempted to explore the influence of coronary artery disease and subclinical AS related polymorphisms on the risk of AS in RA, yet most of these studies draw negative results. However, Kisiel et al. reported that similar genetic factors played a pathogenic role in RA patients with early subclinical AS. But the role might decrease with the prolongation of the AS course, which might be resulted from the effect of anti-rheumatic drugs (83). These inconsistent conclusions remind us that our results also need to be verified in other AS population. To sum up, identified DEGs and hub genes, enriched GO and KEGG pathways, gotten TFs and miRNAs, especially suggested candidate drugs could become therapeutic for RA patients with clinical and subclinical AS, which deserved further experimental research and clinical study.
The novelty of our study is mainly featured in following aspects compared with previous researches. Firstly, four study datasets and two validation datasets were used in our study, the results of which were more convincing. Secondly, candidate drugs were identified in our study, which could be therapeutic for patients with RA and AS.
The leading limitation of the current study is its lack of basic and clinical experimental verification. Although we have verified primary results in external datasets, further experiments would provide more powerful evidence for the transformation and application of our findings in clinical work. In addition, though previous studies have reported genetic associations of AS plaques with RA (84,85), enrichment of common genetic factors for RA and AS in these DEGs requires verification.
In conclusion, our comprehensive bioinformatics analyses of transcriptional profiling provides a novel view regarding the potential pathogenesis of AS in RA. Furthermore, exploitation of candidate drugs targeting hub genes might hold great promise in the future fight against the comorbidities.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-4934/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4934/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Castañeda S, Nurmohamed MT, González-Gay MA. Cardiovascular disease in inflammatory rheumatic diseases. Best Pract Res Clin Rheumatol 2016;30:851-69. [Crossref] [PubMed]
- Nurmohamed MT, Heslinga M, Kitas GD. Cardiovascular comorbidity in rheumatic diseases. Nat Rev Rheumatol 2015;11:693-704. [Crossref] [PubMed]
- Fransen J, Kazemi-Bajestani SM, Bredie SJ, et al. Rheumatoid Arthritis Disadvantages Younger Patients for Cardiovascular Diseases: A Meta-Analysis. PLoS One 2016;11:e0157360. [Crossref] [PubMed]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207-10. [Crossref] [PubMed]
- Clough E, Barrett T. The Gene Expression Omnibus Database. Methods Mol Biol 2016;1418:93-110. [Crossref] [PubMed]
- Gautier L, Cope L, Bolstad BM, et al. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20:307-15. [Crossref] [PubMed]
- Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 2013;41:D991-5. [Crossref] [PubMed]
- Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007;23:1846-7. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:2847-9. [Crossref] [PubMed]
- The Gene Ontology Consortium. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res 2019;47:D330-8. [Crossref] [PubMed]
- Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353-61. [Crossref] [PubMed]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27-30. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Doms A, Schroeder M. GoPubMed: exploring PubMed with the Gene Ontology. Nucleic Acids Res 2005;33:W783-6. [Crossref] [PubMed]
- Ben-Hur A, Noble WS. Kernel methods for predicting protein-protein interactions. Bioinformatics 2005;21:i38-46. [Crossref] [PubMed]
- Ewing RM, Chu P, Elisma F, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol 2007;3:89. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447-52. [Crossref] [PubMed]
- Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res 2011;39:D561-8. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8:S11. [Crossref] [PubMed]
- Xia J, Gill EE, Hancock RE. NetworkAnalyst for statistical, visual and network-based meta-analysis of gene expression data. Nat Protoc 2015;10:823-44. [Crossref] [PubMed]
- Zhou G, Soufan O, Ewald J, et al. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Res 2019;47:W234-41. [Crossref] [PubMed]
- Liu ZP, Wu C, Miao H, et al. RegNetwork: an integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database (Oxford) 2015;2015:bav095. [Crossref] [PubMed]
- Chen EY, Tan CM, Kou Y, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 2013;14:128. [Crossref] [PubMed]
- Wu H, Huang J, Zhong Y, et al. DrugSig: A resource for computational drug repositioning utilizing gene expression signatures. PLoS One 2017;12:e0177743. [Crossref] [PubMed]
- Lin YJ, Anzaghe M, Schülke S. Update on the Pathomechanism, Diagnosis, and Treatment Options for Rheumatoid Arthritis. Cells 2020;9:880. [Crossref] [PubMed]
- Zhu Y, Xian X, Wang Z, et al. Research Progress on the Relationship between Atherosclerosis and Inflammation. Biomolecules 2018;8:80. [Crossref] [PubMed]
- Wolf D, Ley K. Immunity and Inflammation in Atherosclerosis. Circ Res 2019;124:315-27. [Crossref] [PubMed]
- Smolen JS, Aletaha D, McInnes IB. Rheumatoid arthritis. Lancet 2016;388:2023-38. [Crossref] [PubMed]
- Tedgui A, Mallat Z. Cytokines in atherosclerosis: pathogenic and regulatory pathways. Physiol Rev 2006;86:515-81. [Crossref] [PubMed]
- Zhao Q, Zhou D, You H, et al. IFN-γ aggravates neointimal hyperplasia by inducing endoplasmic reticulum stress and apoptosis in macrophages by promoting ubiquitin-dependent liver X receptor-α degradation. FASEB J 2017;31:5321-31. [Crossref] [PubMed]
- Dong M, Zhang Y, Xu C, et al. Interferon-γ decreases ATP-binding cassette subfamily G member 1-mediated cholesterol efflux through small ubiquitin-like modifier/ubiquitin-dependent liver X receptor-α degradation in macrophages. Biotechnol Appl Biochem 2021;68:1412-20. [PubMed]
- Elshabrawy HA, Chen Z, Volin MV, et al. The pathogenic role of angiogenesis in rheumatoid arthritis. Angiogenesis 2015;18:433-48. [Crossref] [PubMed]
- Shagdarsuren E, Djalali-Talab Y, Aurrand-Lions M, et al. Importance of junctional adhesion molecule-C for neointimal hyperplasia and monocyte recruitment in atherosclerosis-prone mice-brief report. Arterioscler Thromb Vasc Biol 2009;29:1161-3. [Crossref] [PubMed]
- Keiper T, Al-Fakhri N, Chavakis E, et al. The role of junctional adhesion molecule-C (JAM-C) in oxidized LDL-mediated leukocyte recruitment. FASEB J 2005;19:2078-80. [Crossref] [PubMed]
- Bradfield PF, Menon A, Miljkovic-Licina M, et al. Divergent JAM-C Expression Accelerates Monocyte-Derived Cell Exit from Atherosclerotic Plaques. PLoS One 2016;11:e0159679. [Crossref] [PubMed]
- Huang Q, Pope RM. Toll-like receptor signaling: a potential link among rheumatoid arthritis, systemic lupus, and atherosclerosis. J Leukoc Biol 2010;88:253-62. [Crossref] [PubMed]
- Bourque J, Hawiger D. Immunomodulatory Bonds of the Partnership between Dendritic Cells and T Cells. Crit Rev Immunol 2018;38:379-401. [Crossref] [PubMed]
- Gardner JK, Cornwall SMJ, Musk AW, et al. Elderly dendritic cells respond to LPS/IFN-γ and CD40L stimulation despite incomplete maturation. PLoS One 2018;13:e0195313. [Crossref] [PubMed]
- Zaguri R, Verbovetski I, Atallah M, et al. 'Danger' effect of low-density lipoprotein (LDL) and oxidized LDL on human immature dendritic cells. Clin Exp Immunol 2007;149:543-52. [Crossref] [PubMed]
- Xiao Y, Shi M, Qiu Q, et al. Piperlongumine Suppresses Dendritic Cell Maturation by Reducing Production of Reactive Oxygen Species and Has Therapeutic Potential for Rheumatoid Arthritis. J Immunol 2016;196:4925-34. [Crossref] [PubMed]
- Gunter S, Solomon A, Tsang L, et al. Apelin concentrations are associated with altered atherosclerotic plaque stability mediator levels and atherosclerosis in rheumatoid arthritis. Atherosclerosis 2017;256:75-81. [Crossref] [PubMed]
- Ram M, Sherer Y, Shoenfeld Y. Matrix metalloproteinase-9 and autoimmune diseases. J Clin Immunol 2006;26:299-307. [Crossref] [PubMed]
- Ibrahem EM, El-Gendi SS, Mahmoud AA, et al. Predictors of Cardiovascular Affection in Patients with Active Rheumatoid Arthritis: Secondary Analysis of a Randomized Controlled Trial. Curr Rheumatol Rev 2021;17:258-66. [Crossref] [PubMed]
- Otto M, Dorn B, Grasmik T, et al. Apremilast effectively inhibits TNFα-induced vascular inflammation in human endothelial cells. J Eur Acad Dermatol Venereol 2022;36:237-46. [Crossref] [PubMed]
- Rudra DS, Pal U, Maiti NC, et al. Melatonin inhibits matrix metalloproteinase-9 activity by binding to its active site. J Pineal Res 2013;54:398-405. [Crossref] [PubMed]
- Crump KE, Sahingur SE. Microbial Nucleic Acid Sensing in Oral and Systemic Diseases. J Dent Res 2016;95:17-25. [Crossref] [PubMed]
- Lim SY, Raftery MJ, Goyette J, et al. Oxidative modifications of S100 proteins: functional regulation by redox. J Leukoc Biol 2009;86:577-87. [Crossref] [PubMed]
- Abreu JR, Krausz S, Dontje W, et al. Sustained T cell Rap1 signaling is protective in the collagen-induced arthritis model of rheumatoid arthritis. Arthritis Rheum 2010;62:3289-99. [Crossref] [PubMed]
- Kojima F, Kapoor M, Kawai S, et al. Prostaglandin E2 activates Rap1 via EP2/EP4 receptors and cAMP-signaling in rheumatoid synovial fibroblasts: involvement of Epac1 and PKA. Prostaglandins Other Lipid Mediat 2009;89:26-33. [Crossref] [PubMed]
- Remans PH, Wijbrandts CA, Sanders ME, et al. CTLA-4IG suppresses reactive oxygen species by preventing synovial adherent cell-induced inactivation of Rap1, a Ras family GTPASE mediator of oxidative stress in rheumatoid arthritis T cells. Arthritis Rheum 2006;54:3135-43. [Crossref] [PubMed]
- Singh B, Kosuru R, Lakshmikanthan S, et al. Endothelial Rap1 (Ras-Association Proximate 1) Restricts Inflammatory Signaling to Protect From the Progression of Atherosclerosis. Arterioscler Thromb Vasc Biol 2021;41:638-50. [Crossref] [PubMed]
- Cai Y, Sukhova GK, Wong HK, et al. Rap1 induces cytokine production in pro-inflammatory macrophages through NFκB signaling and is highly expressed in human atherosclerotic lesions. Cell Cycle 2015;14:3580-92. [Crossref] [PubMed]
- Bretones G, Delgado MD, León J. Myc and cell cycle control. Biochim Biophys Acta 2015;1849:506-16. [Crossref] [PubMed]
- Lee YZ, Guo HC, Zhao GH, et al. Tylophorine-based compounds are therapeutic in rheumatoid arthritis by targeting the caprin-1 ribonucleoprotein complex and inhibiting expression of associated c-Myc and HIF-1α. Pharmacol Res 2020;152:104581. [Crossref] [PubMed]
- Pap T, Nawrath M, Heinrich J, et al. Cooperation of Ras- and c-Myc-dependent pathways in regulating the growth and invasiveness of synovial fibroblasts in rheumatoid arthritis. Arthritis Rheum 2004;50:2794-802. [Crossref] [PubMed]
- Xu L, Hao H, Hao Y, et al. Aberrant MFN2 transcription facilitates homocysteine-induced VSMCs proliferation via the increased binding of c-Myc to DNMT1 in atherosclerosis. J Cell Mol Med 2019;23:4611-26. [Crossref] [PubMed]
- Khanna A. Concerted effect of transforming growth factor-beta, cyclin inhibitor p21, and c-myc on smooth muscle cell proliferation. Am J Physiol Heart Circ Physiol 2004;286:H1133-40. [Crossref] [PubMed]
- Xiao J, Wang R, Zhou W, et al. LncRNA NEAT1 regulates the proliferation and production of the inflammatory cytokines in rheumatoid arthritis fibroblast-like synoviocytes by targeting miR-204-5p. Hum Cell 2021;34:372-82. [Crossref] [PubMed]
- Chen J, Luo X, Liu M, et al. Silencing long non-coding RNA NEAT1 attenuates rheumatoid arthritis via the MAPK/ERK signalling pathway by downregulating microRNA-129 and microRNA-204. RNA Biol 2021;18:657-68. [Crossref] [PubMed]
- Xu F, Zhong JY, Lin X, et al. Melatonin alleviates vascular calcification and ageing through exosomal miR-204/miR-211 cluster in a paracrine manner. J Pineal Res 2020;68:e12631. [Crossref] [PubMed]
- Liu X, Guo JW, Lin XC, et al. Macrophage NFATc3 prevents foam cell formation and atherosclerosis: evidence and mechanisms. Eur Heart J 2021;42:4847-61. [Crossref] [PubMed]
- Yu C, Li L, Xie F, et al. LncRNA TUG1 sponges miR-204-5p to promote osteoblast differentiation through upregulating Runx2 in aortic valve calcification. Cardiovasc Res 2018;114:168-79. [Crossref] [PubMed]
- Wang Z, Zhang M, Wang Z, et al. Cyanidin-3-O-glucoside attenuates endothelial cell dysfunction by modulating miR-204-5p/SIRT1-mediated inflammation and apoptosis. Biofactors 2020;46:803-12. [Crossref] [PubMed]
- Ghasempour G, Mohammadi A, Zamani-Garmsiri F, et al. miRNAs through β-ARR2/p-ERK1/2 pathway regulate the VSMC proliferation and migration. Life Sci 2021;279:119703. [Crossref] [PubMed]
- Wang H, Wei Z, Li H, et al. MiR-377-3p inhibits atherosclerosis-associated vascular smooth muscle cell proliferation and migration via targeting neuropilin2. Biosci Rep 2020;40:BSR20193425. [Crossref] [PubMed]
- Guo Y, Huang S, Ma Y, et al. MiR-377 mediates the expression of Syk to attenuate atherosclerosis lesion development in ApoE-/- mice. Biomed Pharmacother 2019;118:109332. [Crossref] [PubMed]
- Zhang P, Wang W, Li M. Circ_0010283/miR-377-3p/Cyclin D1 Axis Is Associated With Proliferation, Apoptosis, Migration, and Inflammation of Oxidized Low-density Lipoprotein-Stimulated Vascular Smooth Muscle Cells. J Cardiovasc Pharmacol 2021;78:437-47. [Crossref] [PubMed]
- Chen LY, Xia XD, Zhao ZW, et al. MicroRNA-377 Inhibits Atherosclerosis by Regulating Triglyceride Metabolism Through the DNA Methyltransferase 1 in Apolipoprotein E-Knockout Mice. Circ J 2018;82:2861-71. [Crossref] [PubMed]
- Niu M, Li H, Li X, et al. Circulating Exosomal miRNAs as Novel Biomarkers Perform Superior Diagnostic Efficiency Compared With Plasma miRNAs for Large-Artery Atherosclerosis Stroke. Front Pharmacol 2021;12:791644. [Crossref] [PubMed]
- Chang TK, Zhong YH, Liu SC, et al. Apelin Promotes Endothelial Progenitor Cell Angiogenesis in Rheumatoid Arthritis Disease via the miR-525-5p/Angiopoietin-1 Pathway. Front Immunol 2021;12:737990. [Crossref] [PubMed]
- Strand KA, Lu S, Mutryn MF, et al. High Throughput Screen Identifies the DNMT1 (DNA Methyltransferase-1) Inhibitor, 5-Azacytidine, as a Potent Inducer of PTEN (Phosphatase and Tensin Homolog): Central Role for PTEN in 5-Azacytidine Protection Against Pathological Vascular Remodeling. Arterioscler Thromb Vasc Biol 2020;40:1854-69. [Crossref] [PubMed]
- Dunn J, Simmons R, Thabet S, et al. The role of epigenetics in the endothelial cell shear stress response and atherosclerosis. Int J Biochem Cell Biol 2015;67:167-76. [Crossref] [PubMed]
- Cao Q, Wang X, Jia L, et al. Inhibiting DNA Methylation by 5-Aza-2'-deoxycytidine ameliorates atherosclerosis through suppressing macrophage inflammation. Endocrinology 2014;155:4925-38. [Crossref] [PubMed]
- Tóth DM, Ocskó T, Balog A, et al. Amelioration of Autoimmune Arthritis in Mice Treated With the DNA Methyltransferase Inhibitor 5'-Azacytidine. Arthritis Rheumatol 2019;71:1265-75. [Crossref] [PubMed]
- Li XF, Wu S, Yan Q, et al. PTEN Methylation Promotes Inflammation and Activation of Fibroblast-Like Synoviocytes in Rheumatoid Arthritis. Front Pharmacol 2021;12:700373. [Crossref] [PubMed]
- Mosquera N, Rodriguez-Trillo A, Blanco FJ, et al. All-Trans Retinoic Acid Inhibits Migration and Invasiveness of Rheumatoid Fibroblast-Like Synoviocytes. J Pharmacol Exp Ther 2020;372:185-92. [Crossref] [PubMed]
- Cui Z, Lin Y, Liu Y, et al. Retinoic Acid-Platinum (II) Complex [RT-Pt(II)] Protects Against Rheumatoid Arthritis in Mice via MEK/Nuclear Factor kappa B (NF-κB) Pathway Downregulation. Med Sci Monit 2020;26:e924787. [Crossref] [PubMed]
- Pan H, Xue C, Auerbach BJ, et al. Single-Cell Genomics Reveals a Novel Cell State During Smooth Muscle Cell Phenotypic Switching and Potential Therapeutic Targets for Atherosclerosis in Mouse and Human. Circulation 2020;142:2060-75. [Crossref] [PubMed]
- Krivospitskaya O, Elmabsout AA, Sundman E, et al. A CYP26B1 polymorphism enhances retinoic acid catabolism and may aggravate atherosclerosis. Mol Med 2012;18:712-8. [Crossref] [PubMed]
- Kisiel B, Kruszewski R, Juszkiewicz A, et al. Common atherosclerosis genetic risk factors and subclinical atherosclerosis in rheumatoid arthritis: the relevance of disease duration. Rheumatol Int. 2019;39:327-36. [Crossref] [PubMed]
- Arya R, Escalante A, Farook VS, et al. A genetic association study of carotid intima-media thickness (CIMT) and plaque in Mexican Americans and European Americans with rheumatoid arthritis. Atherosclerosis 2018;271:92-101. [Crossref] [PubMed]
- López-Mejías R, Genre F, García-Bermúdez M, et al. The ZC3HC1 rs11556924 polymorphism is associated with increased carotid intima-media thickness in patients with rheumatoid arthritis. Arthritis Res Ther 2013;15:R152. [Crossref] [PubMed]
(English Language Editor: B. Draper)


