
Screening of plasma exosomal lncRNAs to identify potential biomarkers for obstructive sleep apnea
Introduction
Obstructive sleep apnea (OSA) is a disease of repeated hypoventilation and apnea caused by upper airway obstruction, further resulting in intermittent hypoxemia and sleep structure disorders, affecting 936 million adults between the ages of 30 to 69 worldwide (1). The main pathophysiological mechanism of OSA is considered to be chronic hypoxia, accompanied by an inflammatory response, oxidative stress, and other factors (2). In recent years, OSA incidence has gradually increased (3). The impacts of OSA include daytime drowsiness, nocturnal wakefulness, memory loss, changes in the respiratory, metabolic, and cardiovascular systems, and neuropsychiatric alterations. Therefore, OSA seriously threatens human health and affects quality of life (4,5). Hypertension (HTN) is a common complication of OSA (6). Studies have shown that more than 70% of refractory HTN patients experience OSA (7). Additionally, OSA is recognized internationally as the cause of secondary HTN; HTN, in turn, might cause or even aggravate OSA (8). The gold standard diagnostic approach for OSA is sleep study through polysomnography (PSG), which is an in-laboratory test, technically demanding and time-consuming, expensive is the core reason for limiting the accessibility. In previous studies, biomarkers of inflammation, oxidative stress, glycolipid metabolism were mainly studied as biomarkers of OSA, but there have been conflicting statements, and the sensitivity and specificity are obviously insufficient. Therefore, finding molecular markers to allow correct identification of OSA with and without HTN and understanding OSA pathogenesis is essential for the early adoption of appropriate therapeutic interventions and for the determination of new therapeutic targets.
Recently, long non-coding RNAs (lncRNAs), which are 200-nucleotide (nt) long transcripts without an open reading frame, have emerged as regulators in many biological processes (9). They have been involved in the regulation of OSA progression (10). For example, overexpression of the lncRNA CPS1-IT reduces pulmonary arterial hypertension in OSA patients by decreasing interleukin-1β (IL-1β) expression through hypoxia inducible factor 1 (HIF1) transcriptional activity (11). In addition, blocking the lncRNA axis metastasis-associated lung adenocarcinoma transcript 1 (MALAT1)/miR-224-5p/NLR family pyrin domain containing 3 (NLRP3) inhibits the hippocampal inflammatory response of patients with type 2 diabetes mellitus and OSA (12). The above evidences indicate that lncRNAs have important functions in the development of OSA and their clinical application value in the future. Therefore, OSA-related lncRNAs represent a new class of diagnostic markers or therapeutic targets.
One of the challenges in developing non-invasive methods for diagnostic analyses of body fluids is the ability to identify clinically relevant biomarkers (13). Exosomes are small, single-membrane secretory organelles of 30–200 nm in diameter involved in intercellular communication, including protein and RNA transmission (14). Exosomes contain a variety of proteins, nucleic acids, and lipids and are present in many if not all body fluids (15). Notably, exosomal lncRNAs exhibit diverse expression patterns in different cells or various physiological and pathological conditions. Many studies have focused on the relationship between exosomes and their content with complex diseases. It has been reported that OSA might initiate and intensify cell senescence through exosomes, possibly through oxidative stress-related pathways (16). Exosomal lncRNAs expressed stably, easily and rapidly measurable, have been reported as novel biomarkers for many diseases. For example, the plasma exosomal lncRNA SOX2-OT might serve as a biomarker for lung squamous cell carcinoma (17); SLC2A12-10:1 has been proposed as a new biomarker for the diagnosis of gastric cancer (18). However, a role for plasma exosomal lncRNAs in OSA has not been reported.
Therefore, the present study explored the differential expression of lncRNAs in plasma exosomes of non-OSA and OSA patients using high-throughput sequencing to identify new biomarkers for the non-invasive diagnosis of OSA as well as new potential molecular targets for its treatment. We present the following article in accordance with the STARD reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3818/rc).
Methods
Collection of clinical samples
First, 38 patients hospitalized in the Nanfang Hospital, Southern Medical University were selected. This was a clinical study design that was rigorously matched to incorporate important demographic factors. They were divided into two groups according to their Apnea-Hypopnea Index (AHI): OSA (AHI ≥5 events/h, n=23) or HTN-OSA group (AHI ≥5; systolic BP ≥140 and/or diastolic BP ≥90 mmHg, n=15); a further 25 individuals with normal blood pressure and without OSA were selected as negative controls (NC group, n=25), and verified using an independent validation with 31 OSA patients and 10 normal cases. According to their AHI, OSA patients were divided into an OSA mild group (5≤ AHI <15 events/h, n=10), an OSA moderate group (15≤ AHI <30 events/h, n=11), and an OSA severe group (AHI ≥30 events/h, n=10). The exclusion criteria were as follows: (I) presence of secondary HTN; (II) diagnosis of diabetes and irregular use of hypoglycemic drugs; (III) acute cardiovascular and cerebrovascular events in the past 3 months; and (IV) presence of chronic respiratory disease, bronchial asthma, or accepted treatment with continuous positive airway pressure (CPAP).
The plasma was collected from all participants as shown in Table 1, and clinical characteristics were recorded. In addition, the clinical characteristics of OSA patients with different severities were also recorded in Table 2. This experiment was approved by the Ethics Committee of Nanfang Hospital of Southern Medical University (approval No. NFEC-2021-436) and written informed consent was signed by each patient. All methods were performed in accordance with current clinical practice guidelines and regulations. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Table 1
| Parameter | NC group (n=25) | OSA group (n=23) | HTN-OSA group (n=15) |
|---|---|---|---|
| Age (years) | 38.92±11.31 | 47.30±12.50* | 51.53±11.63** |
| Gender (male, %) | 52% | 52% | 60% |
| BMI (kg/m2) | 22.13±1.68 | 23.86±3.70 | 25.71±3.13**† |
| SBP (mm Hg) | 114.8±8.90 | 120.65±10.02 | 144.93±21.55**†† |
| DBP (mm Hg) | 72.04±7.79 | 81.39±12.34** | 93.27±15.13**†† |
| AHI (events/h) | 2.43±1.25 | 10.16±14.78** | 13.43±26.18** |
| LSaO2 (%) | 90.12±2.70 | 81.65±11.96** | 80.33±10.23** |
All data were expressed as mean ± standard deviation. *P<0.05, **P<0.01, OSA group or HTN-OSA group versus NC group; †P<0.05, ††P<0.01, HTN-OSA group versus OSA group. BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure; AHI, apnea and hypopnea index; LSaO2, lowest oxygen saturation; NC, normal control; OSA, obstructive sleep apnea; HTN, hypertension.
Table 2
| Parameter | NC group (n=10) | OSA mild group (n=10) | OSA moderate group (n=11) | OSA severe group (n=10) |
|---|---|---|---|---|
| Age (years) | 36.80±13.21 | 50.80±12.16 | 43.55±15.27 | 40.50±9.28 |
| BMI (kg/m2) | 22.28±1.40 | 23.18±1.97 | 26.19±2.53** | 27.35±5.02** |
| Neck circumference (cm) | 37.30±3.62 | 34.80±4.23 | 38.23±2.52 | 38.85±3.71 |
| Friedman | 1.20±0.42 | 2.10±1.20 | 2.91±0.83** | 2.60±0.84* |
| AHI (events/h) | 2.24±0.94 | 5.62±2.37 | 22.59±2.84** | 54.42±15.53**†† |
| ODI (events/h) | 1.37±0.83 | 4.27±2.94 | 17.08±8.29* | 51.29±26.31**†† |
| TS90 (%) | 1.13±3.15667 | 0.114±0.18992 | 2.2364±1.0312†† | 27.54±24.61**†† |
| LSaO2 (%) | 89.80±2.94 | 89.20±2.20 | 80.55±5.09**† | 66.30±17.24**†† |
| MSaO2 (%) | 94.30±1.25 | 95.00±1.70 | 94.55±1.21 | 91.60±2.91† |
All data were expressed as mean ± standard deviation. *P<0.05, **P<0.01, NC verse other groups; †P<0.05, ††P<0.01, OSA mild group verse OSA medium group or OSA severe group. BMI, body mass index; NC, normal control; AHI, apnea hypopnea index; ODI, oxygen desaturation index; LSaO2, lowest oxygen saturation; MSaO2, mean oxygen saturation; OSA, obstructive sleep apnea.
Exosome extraction
Exosomes were isolated and extracted using the ultracentrifugation method. First, plasma samples were rapidly thawed at 37 ℃, transferred into centrifugation tubes, and centrifuged at 2,000 g for 30 minutes at 4 ℃. Supernatants were carefully transferred into new centrifugation tubes and centrifuged at 10,000 g for 45 minutes at 4 ℃ to remove larger vesicles. Then, supernatants were filtered through a 0.45 µm filter membrane. After filtration, samples were transferred into centrifugation tubes and centrifuged at 100,000 g for 70 minutes at 4 ℃. Pellets were resuspended with 10 mL of precooled 1× phosphate-buffered saline (PBS) and centrifuged at 100,000 g for 70 minutes at 4 ℃. Finally, pellets were resuspended with 200–300 µL of precooled 1× PBS to obtain the exosomes.
Transmission electron microscopy
Exosomes (10 µL) were added to the copper grid for 1 minute and the remaining liquid was absorbed with filter paper. Then, 10 µL uranyl acetate was added for 1 minute and the remaining liquid was absorbed with filter paper. Grids were dried for several minutes at room temperature and observed under a transmission electron microscope (TEM; HT-7700; Hitachi, Tokyo, Japan) at 100 kv.
Nanoparticle tracking analysis
Exosome samples were diluted with 1× PBS and directly used for nanoparticle tracking analysis (NTA; ZetaView; Particle Metrix, Ammersee, Germany). According to the minimum expected particle radius and track length set by the system, the diluted samples were recorded 3 times at the speed of 30 frames per second. The resulting complete track measurements were analyzed.
Protein extraction and concentration determination from exosomes
The exosomes were rapidly thawed at 37 ℃ and quickly mixed with 6× radioimmunoprecipitation assay (RIPA) lysis buffer. The solution was then placed on ice for 30 minutes with regular mixing. The standards used for protein concentration determination were prepared according to the bicinchoninic acid (BCA) method. To measure the protein concentration, 5 µL of samples were added to the BCA mixture and mixed well. After incubation at 37 ℃ for 30 minutes, absorbance values at optical density (OD) 562 nm were measured on a microplate reader (Varioskan LUX; Thermo Fisher Scientific, Waltham, MA, USA). The sample protein concentrations were determined using the standard curve.
Western blot analysis of exosomal markers
After determination of the protein concentration, samples were mixed with loading buffer and boiled in a water bath for 5 minutes. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed and the proteins were transferred onto polyvinylidene fluoride (PVDF) membrane. The membranes were blocked with 5% skim milk solution at room temperature for 2 hours and incubated with primary antibodies recognizing CD9 (AB92726, Abcam, Cambridge, MA, USA), CD63 (A5271, Abclonal, Wuhan, China), TSG101 (AB125011, Abcam), and calnexin (AB22595, Abcam) overnight at 4 ℃. Afterward, membranes were incubated with secondary antibodies and then with enhanced chemiluminescence (ECL). Protein bands were detected using chemiluminescence imaging system (ChemiScope 3000mini; Clinx Science Instruments, Shanghai, China).
Sequencing of lncRNAs and bioinformatics analysis of exosomes
First, a lncRNA database was constructed. Briefly, after extraction of total RNAs, ribosomal RNAs (rRNAs) were removed using the Ribo-Zero kit (MRZG12324; Epicenter, Madison, WI, USA), followed by RNA interruption, double-stranded complementary DNA (cDNA) synthesis, terminal repair, addition of polyA tail, and splicing. Then, we selected fragments for polymerase chain reaction (PCR) amplification and purification. Finally, the library quality was assessed, and the sequencing was performed. StringTie software (https://ccb.jhu.edu/software/stringtie/) was used to construct transcripts using comparison files. Annotated and candidate lncRNA transcripts were obtained by comparing with human genome annotation files using gFFCompare in StringTie. Transcripts of length >200 nt were selected as candidate lncRNA transcripts. We then used 4 kinds of software, namely, CPC2 (https://github.com/gao-lab/CPC2_standalone), CNCI (https://github.com/www-bioinfo-org/CNCI), Pfam (Pfamscan: https://www.ebi.ac.uk/Tools/pfa/pfamscan/) and FEElnc (FEElnc: https://github.com/tderrien/FEELnc), to predict the coding ability of the lncRNAs. At least 3 kinds of software were chosen to predict the coding transcripts and obtain new lncRNAs. Differential gene expression was analyzed using DESeq2 software (https://bioconductor.org/packages/release/bioc/html/DESeq2.html), and differentially expressed lncRNsA (known and unknown) were quantified and annotated using featureCounts (http://subread.sourceforge.net/). The volcanic map of gene expression was drawn with the Log2 of the fold change as abscissa and -Log10 of the P-value as ordinate. Pair-differentially expressed lncRNAs were subjected to Venn analysis. The messenger RNAs (mRNAs) were quantified and annotated on the comparison file using featureCounts. The DESeq2 software was used to analyze the differential mRNA expression. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was used to identify the most represented biochemical metabolic and signaling pathways among the genes. Adjusted P value (Padj) was calculated with the Benjamini-Hochberg method. The first 15 pathways with the smallest Padj were selected for the bubble map. In addition, cis-target gene prediction suggested that the function of lncRNAs was related to the mRNAs adjacent to the coordinate. The mRNAs adjacent to lncRNAs (100 kb upstream and downstream) were screened out, and an intersection analyses between each lncRNA and the differentially expressed mRNAs found 100 kb upstream and downstream of this lncRNA (i.e., lncRNA target genes) were performed. Subsequently, the main functions of lncRNAs were predicted by functional enrichment analysis of these target genes. In addition, miRanda (version 3.3; https://bioweb.pasteur.fr/packages/pack@miRanda@3.3) was used to predict the binding sites of plasma exosomal ENST00000592016-regulated micro RNAs (miRNAs) interacting with target genes. The top 10 genes in the relative 3'untranslated region (3'UTR) of each miRNA were selected as candidate target genes. The screened target genes were used for KEGG analysis (http://kobas.cbi.pku.edu.cn/kobas3/?t=1); the OSA-related items reported in the top 20 significant KEGG items were screened, and the pathways associated with OSA were selected.
Droplet digital PCR
We selected lncRNAs differentially expressed in both OSA and HTN-OSA groups when compared to the levels found in the NC group. The screening length was <3,000 bp and the lncRNA type was known. The lncRNAs with absolute values of OSA_NC_log2FC, HTN_NC_log2FC, and HTN_OSA_log2FC greater than 1 were candidate molecules. The expression changes of plasma exosomal ENST00000442889, ENST00000592016, and ENST00000319701 were measured. Next, we detected ENST00000592016 expression in OSA patients with different severity. In brief, cDNA was synthesized using droplet digital PCR (ddPCR) EvaGreen Supermix (Bio-Rad, Hercules, CA, USA) and ddPCR was performed with a QX200 droplet system. The droplets used for PCR reactions were generated according to the manufacturer’s protocol (Bio-Rad). Then, samples were transferred into a 96-well PCR plate for ddPCR (19). The primer sequences were as follows: H-ENST00000319701-Forward: ACAGTGGAGTGAAGCAGACC, H-ENST00000319701-Reverse: TGGCTATCGGCTTCTCATCC; H-ENST00000442889-Forward: GAACGAGCCTAGGAACCGG, H-ENST00000442889-Reverse: GCCCAGGAGCAGAATTCCTT; H-ENST00000592016-Forward: CTCCCTCAAGCGCCTGATC, H-ENST00000592016-Reverse: CTGTGGCACAGCTTCTCTCC.
Statistical analyses
The software GraphPad 8.0 (GraphPad Software Inc., San Diego, CA, USA) was used for statistical analyses. Data were expressed as means ± standard deviation (SD). Comparability was assessed using the Mann-Whitney U test for quantitative characteristics and Fisher’s exact test for qualitative variables. Student’s t-test was used to analyze the differences between two groups and one-way analysis of variance (ANOVA) for comparisons among multiple groups. Receiver operating characteristic (ROC) curve was used to analyze the specificity and sensitivity of exosomal lncRNA ENST00000592016 as a marker for OSA diagnosis. Kruskal-Wallis rank sum test was used to analyze the indicators that did not conform to the normal distribution. Spearman’s rank correlation coefficient was used to evaluate the association between ENST00000592016 levels and polysomnography (PSG) parameters. A P value <0.05 was considered statistically significant.
Results
Isolation and identification of exosomes
First, we briefly analyzed the clinical information of the participants in the first phase, and most of the patients were over middle age. There were significant differences in the baseline characteristics of age and body mass index (BMI) between the HTN-OSA and NC groups, with age (51.53±11.63 vs. 38.92±11.31 years) and BMI (25.71±3.13 vs. 22.13±1.68 kg/m2).
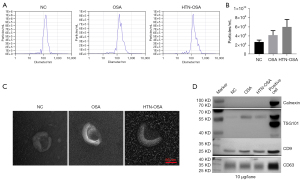
Next, we extracted exosomes from the plasma of the NC, OSA, and HTN-OSA groups. NTA detection showed that the average size of plasma exosomes was about 150 nm in each group (Figure 1A). Exosome particle concentrations were higher in the OSA and HTN-OSA groups than that in the NC group (Figure 1B). The TEM revealed cup-shaped exosomes with a size between 30 and 200 nm, which was consistent with typical exosomes (Figure 1C). Western blot analyses showed that exosome samples contained the typical protein makers TSG101, CD9, and CD63. In contrast, the negative marker calnexin was not expressed in exosomes but was found in positive controls (Hela cells) (Figure 1D). These results confirmed the successful isolation of plasma exosomes.
Analysis of lncRNA expression in exosomes
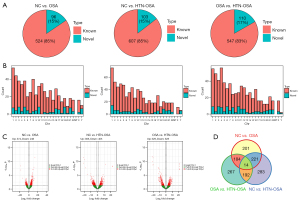
Next, we identified the lncRNAs differentially expressed in isolated exosomes from the three groups. For the P<0.05 and |log2FC| ≥1 conditions, 620 lncRNAs, among which 524 were known and 96 were newly identified, were differentially expressed between the NC and OSA groups. There were 710 lncRNAs, among which 607 were known and 103 were newly identified, differentially expressed between the NC and HTN-OSA groups. Finally, 657 lncRNAs, among which 547 were known and 110 were newly identified, were differentially expressed between the OSA and HTN-OSA group (Figure 2A). Statistical analyses of the chromosomal distribution of the differentially expressed lncRNAs showed that lncRNAs were found on every chromosome but not on mitochondrial DNA (Figure 2B). The volcano plot of differentially expressed lncRNAs showed that under the screening conditions of P<0.05 and |log2FC| ≥1, 374 lncRNAs were upregulated and 246 were downregulated in the OSA group compared with the expression levels in the NC group. In the HTN-OSA group, 305 lncRNAs were upregulated and 405 were downregulated compared with the levels in the NC group. In the OSA group, 329 lncRNAs were upregulated and 328 were downregulated compared with those of the HTN-OSA group (Figure 2C). Venn analysis of paired lncRNAs revealed 14 lncRNAs differentially expressed among the three groups (Figure 2D).
Analysis of mRNA expression in exosomes
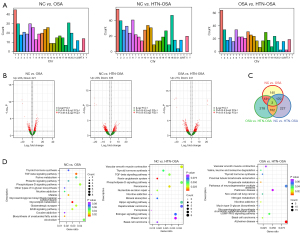
The mRNAs differentially expressed between the NC and OSA groups and the OSA and HTN-OSA group were mainly found on chromosomes other than Y and in mitochondria. The mRNAs differentially expressed between the NC and HTN-OSA group were distributed on all chromosomes except Y (Figure 3A). The volcano plot showed that under the screening conditions P<0.05 and |log2FC| ≥1, 444 mRNAs were differentially expressed between the NC and OSA groups, 562 between the NC and HTN-OSA groups, and 514 between the OSA and HTN-OSA groups (Figure 3B). Venn analysis was performed on pair mRNAs, and identified 3 mRNAs differentially expressed among the three groups (Figure 3C). In addition, the KEGG enrichment analysis of the differentially expressed mRNAs showed that most of the enriched pathways had been linked to OSA (Figure 3D). Among them, the transforming growth factor-β (TGF-β) pathway (20), glutamatergic synapse (21), and ERBB pathway (22) have been reported to be closely relevant to OSA.
Cis-target analysis of differentially expressed exosomal lncRNAs
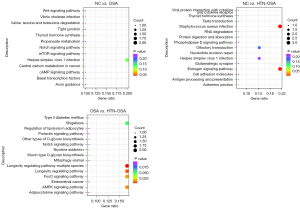
Cis-target gene prediction suggested that the functions of a lncRNA were related to the mRNAs adjacent to its coordinate. We investigated the lncRNA main functions by functional enrichment analysis of target genes. Figure 4 shows the KEGG enrichment analysis of the 15 most differentially expressed lncRNAs. Most of the enriched pathways had been linked to OSA, including the mammalian target of rapamycin (mTOR) pathway (23), the WNT signaling pathway (24), and the glutamatergic synapse pathway (21), among others.
Detection of differentially expressed lncRNAs in exosomes
Next, lncRNAs candidates that were differentially expressed between NC and OSA patients, NC and HTN-OSA patients, and OSA and HTN-OSA patients were identified. We selected 3 lncRNAs from the following for verification based on their length and fold change: The screening length was <3,000 bp and the lncRNAs type were known. OSA_NC_log2FC, HTN_NC_log2FC, and HTN_OSA_log2FC lncRNAs with absolute values greater than 1. Table 3 shows the differentially expressed exosomal lncRNAs.
Table 3
| Gene ID | Length | Type | OSA_NC_log2FC | OSA_NC_p | HTN_NC_log2FC | HTN_NC_p | HTN_OSA_log2FC | HTN_OSA_p |
|---|---|---|---|---|---|---|---|---|
| ENST00000319701 | 2,217 | Known | −6.944371697 | 0.01781369 | -8.414881157 | 0.003950747 | −1.47050946 | 0.639466069 |
| ENST00000561588 | 1,050 | Known | −6.720376967 | 0.025848975 | −8.128366982 | 0.006773451 | −1.407990015 | 0.661737508 |
| ENST00000567491 | 1,604 | Known | −6.49279114 | 0.029912854 | −7.901550508 | 0.007937775 | −1.408759368 | 0.659177954 |
| ENST00000442889 | 952 | Known | −6.009406532 | 0.020251904 | −7.432505745 | 0.003867084 | −1.423099213 | 0.61390071 |
| ENST00000592016 | 2,692 | Known | −6.000381949 | 0.045400987 | −7.46982014 | 0.012421646 | −1.469438192 | 0.646218779 |
| ENST00000662488 | 2,827 | Known | −5.876933956 | 0.049338482 | −7.858074074 | 0.008844651 | −1.981140118 | 0.535018746 |
| ENST00000594590 | 2,179 | Known | −5.432287707 | 0.029991705 | −6.476888889 | 0.009467524 | −1.044601182 | 0.698299424 |
lncRNA, long non-coding RNA; OSA, obstructive sleep apnea.
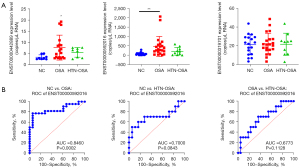
As shown in Figure 5A, ddPCR detected expression changes of exosomal ENST00000442889, ENST00000592016, and ENST00000319701 in the plasma. The expression of ENST00000442889 was low and was not detected in most samples. The expression of ENST00000592016 was higher in the OSA group than that in the NC group, whereas it was not significantly affected in the HTN-OSA group. The levels of ENST00000592016 in the HTN-OSA group were slightly decreased compared with those in the OSA group, but the difference was not statistically significant. The expression of ENST00000319701 was moderate, and not significantly different among groups. The ROC curves were further used to analyze the specificity and sensitivity of ENST00000592016 as OSA diagnostic marker. Figure 5B showed the different comparisons: area under the curve (AUC) =0.8460 (95% CI: 0.72–0.97, P=0.0002) for NC versus OSA and the sensitivity and specificity for diagnosing OSA patients at the best cut-off value (71.71) were 77.27% and 94.4%, respectively. AUC =0.7000 (95% CI: 0.47–0.93, P=0.0843) for NC versus HTN-OSA, and AUC =0.6773 (95% CI: 0.48–0.88, P=0.1128) for OSA versus HTN-OSA. These results indicated that ENST00000592016 expression allowed to NC groups from OSA samples but did not distinguish NC from HTN-OSA or OSA from HTN-OSA conditions.
ENST00000592016 changes with different severity of OSA patients and its correlation with PSG parameter
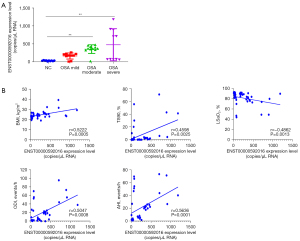
We collected plasma exosomes from 10 normal individuals and 31 OSA patients, and further analyzed the differential expression of the screened ENST00000592016 in OSA patients with different severity. Table 2 shows the general data characteristics of NC and OSA of different severity. Among them, compared with NC group, BMI, AHI, oxygen desaturation index (ODI) increased and lowest oxygen saturation (LSaO2) decreased in the moderate and severe group, and those of the mild group showed no obvious change. In addition, the ddPCR results showed that ENST00000592016 expression in the OSA group was significantly higher than that in the NC group, and the expression gradually increased from mild to severe (Figure 6A). Then, we used Spearman analysis to analyze the correlation between ENST00000592016 expression and clinical characteristics. As shown in Figure 6B, ENST00000592016 expression was positively correlated with BMI, AHI, ODI, and TS90. However, ENST00000592016 expression was negatively correlated with LSaO2.
Interaction network prediction of plasma exosomal ENST00000592016
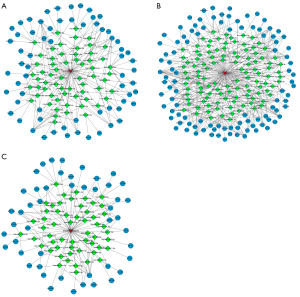
Finally, we used miRanda to predict the possible competing endogenous RNA (ceRNA) interaction network of plasma exosomal ENST00000592016, and then performed KEGG analysis on screened target genes. After reviewing the literature, it was found that the PI3K-Akt, MAPK, and tumor necrosis factor (TNF) signaling pathways were related to OSA. Then, interaction pairs targeting these 3 pathways were selected to draw the interaction network map respectively. Figure 7 shows the lncRNA-miRNA-mRNA regulatory network with ENST00000592016 as the core, mainly involving the PI3K-Akt (Figure 7A), MAPK (Figure 7B), and TNF signaling pathways (Figure 7C).
Discussion
Although OSA is an independent risk factor for HTN, both are thought to increase the risk of cardiovascular events and all-cause death. Untreated OSA has been linked to an increased risk of HTN and HTN might increase the risk of OSA (25,26). Due to the insidious nature and heterogeneity of OSA, it is crucial to identify a molecular marker that can reflect the characteristics of early OSA occurrence. Therefore, finding molecular markers which enable correct identification of OSA, even HTN-OSA, is essential for the early adoption of appropriate therapeutic interventions. The present study was the first to explore the differential expression of lncRNAs in plasma exosomes of non-OSA and OSA patients with or without HTN by high-throughput sequencing to identify potential biomarkers for non-invasive diagnosis of OSA and HTN-OSA. This is also the first report identifying a plasma exosomal lncRNA as a biomarker for OSA.
Exosomes are endogenous lipid bilayer vesicles containing specific nucleic acids, such as DNA, mRNAs, miRNAs, and lncRNAs. Exosomes have been implicated as involved in cell-cell communication (27,28). Exosomal nucleic acids have great potential as disease biomarkers. For example, lncRNAs from plasma exosomes are used as biomarkers in many diseases, including lung squamous cell carcinoma (17), gastric cancer (18), colorectal cancer (29), and Alzheimer’s disease (30). However, the expression characteristics and biological role of plasma exosomal lncRNA in OSA remain unclear. In the present study, we isolated plasma exosomes from normal controls and OSA patients with or without HTN and analyzed then by high-throughput sequencing. We found that lncRNAs and mRNAs were differentially expressed in plasma exosomes of the NC, OSA, and HTN-OSA groups. Screening these lncRNAs might provide biomarkers for OSA.
Enrichment or over expression analyses are commonly used in bioinformatics studies of transcriptome, metabolomics, and microbiome datasets (31). In organisms, different genes coordinate their actions to exercise their biological functions, and pathway-based analysis helps to further understand the biological functions of genes (32). The KEGG is the major public database regarding pathways (33). Pathway enrichment analysis has been conducted using KEGG to determine which pathways were significantly enriched in specific genes compared with the whole genome by using super-hypothesis test. Here, based on the KEGG pathways enriched in differentially expressed mRNAs, we discovered a large number of potential enrichment pathways, which might be associated with OSA. The TGF-β is a multifunctional peptide regulating cell proliferation and differentiation, which is relevant to intimal hyperplasia and vascular remodeling (34). Multiple studies have confirmed that TGF-β can participate in OSA development (35,36). The reason might be that under the condition of chronic inflammation and hypoxia, plentiful inflammatory mediators will be deposited in the subcutaneous layer, induce the synthesis and release of serum TGF-β in OSA patients through a series of signals. Glutamate is one of the most abundant excitatory neurotransmitters in the central nervous system (37). Research has shown that glutamate can also be involved in the occurrence and maintenance of sleep and waking, and its role is to cause and maintain the excitatory state of the cortex, and participate in the regulation of muscle tension during sleep (38,39). The development of glutamatergic synapse has also been reported to be associated with the release of neurotransmitter (40). Studies have also confirmed that neurotransmitters play a key role in sleep-wake system in OSA patients (41,42). As a signaling pathway in brain neurons, ErbB has a protective effect on neurons (43). Research also showed that Nrdp1 might affect the cognition deficits associated with OSA by regulating ErbB (44). Therefore, we preliminarily speculated that the TGF-β, glutamatergic synapse, and ErbB pathways might be the key pathways in the regulation of the OSA process. However, the specific roles and mechanisms of these 3 pathways in OSA are unclear and need further investigation.
Next, we performed cis-target analysis of differentially expressed exosomal lncRNAs. Cis-target gene prediction suggested that the function of a lncRNA was linked to mRNAs adjacent to its coordinate. We discovered that most KEGG pathways enriched in the differentially expressed lncRNAs is related to relevant canonical pathways, mainly containing mTOR (23), WNT (24), and glutamatergic synapse (21).
Autophagy is a survival mechanism for cells to adapt to harsh environments (45). Autophagy the main way to remove abnormal aggregation proteins and damaged organelles in neurons after cerebral ischemia (46,47). Meanwhile, mTOR, is the core gene of the PI3K/Akt/mTOR pathway, and its expression is related to cell cycle and energy metabolism (48). Besides, mTOR also regulates immune response, apoptosis, and autophagy (49). It has been confirmed that the mTOR pathway can participate in the development of OSA-associated HTN (50), in concordance with the hypothesis of a difference in HTN risk between non-OSA and OSA patients. Further, WNT is an evolutionarily conserved pathway that controls cell growth, differentiation, apoptosis, and self-renewal (51). The WNT pathway is crucial in the cognitive impairment caused by OSA (24), These results are in accordance with previous literature in which the impact of OSA on cognitive impairment. Glutamatergic synapse has been introduced in the previous paragraph. Therefore, we concluded that these differentially expressed exosomal lncRNAs are likely to affect autophagy, apoptosis, and glutamate in OSA progression through these pathways, which indicates that they may play a synergistic role in the development of OSA.
Our study suggested that the functional annotations of differentially expressed mRNAs and cis-target genes of differentially expressed lncRNAs were basically consistent, indicating that lncRNAs, as a regulatory factor, play a certain role in the occurrence and development mechanism of OSA. However, the present study is a preliminarily investigation on the role of these lncRNAs and the mechanisms involved need to be further studied.
Direct quantification and clonal amplification of DNA were achieved using ddPCR, which is a biotechnological improvement of the traditional PCR method (52). The application of ddPCR for the detection of different pathogens has rapidly increased because of its ability to accurately detect and quantify low-abundance targets (53). Here, we used ddPCR to detect the differential expression of lncRNAs from plasma exosomes. Due to the low abundance of lncRNAs in exosomes, we focused more on the changing trends. The expression of ENST00000442889 was low and could not be detected in most samples. The expression of ENST00000319701 was moderate and was not significantly different among groups. The expression of ENST00000592016 was high. However, the difference in ENST00000592016 expression measured with ddPCR was not consistent with those obtained by sequencing. The possible reasons for this situation include the differences between the two experimental methods, the small number of sequencing samples and large individual differences. Therefore, the results of large sample verification shall prevail. Our analysis showed that ENST00000592016 expression allowed us to distinguish the NC group from the OSA group, but did not discriminate between the NC and HTN-OSA or the OSA and HTN-OSA groups. These results might be due to the small sample size. Moreover, HTN may affect ENST00000592016 expression preventing the identification of OSA. More research is needed to test this hypothesis.
Studies have shown that moderate-to-severe OSA has previously been shown to be associated with multiple pro-atherogenic mechanisms and increased cardiovascular risk, and the association between OSA and endothelial dysfunction may depend on OSA severity. This suggests that to a certain extent, the clinical features of the disease could reflect the stage of disease development, moderate and severe patients are more prone to develop complications. In our experiment, ENST00000592016 expression also showed a gradually increasing trend from mild, moderate, to severe OSA, which was related to the different severity. Besides, it also showed a good ability to recognize moderate and severe OSA in this study. Whether it is related to the vascular endothelial function caused by the influence of chronic intermittent hypoxia (CIH), while mild OSA fail to cause change of endothelial structure, should be further explore on next research.
To further reveal the possible effect of ENST00000592016 on OSA, the correlation between ENST00000592016 expression and clinical features was analyzed. In a bivariate meta-analysis, Chiu et al. reported age, gender, BMI, study sample size, study population, presence of comorbidities, portable polysomnography (PSG), and bias in index test and reference standard domains risk is an important mediator of OSA sensitivity and specificity (54). In a univariate regression analysis of OSA, gender, BMI, systolic and diastolic blood pressure, AHI, LSaO2, ODI, TS90, and MSaO2 were associated with left ventricular mass index (55). Lisi et al. reported that in controlled HTN, mild-to-moderate OSA may be associated with early diastolic dysfunction independent of age, gender, and mean blood pressure (56). In this study, we found ENST00000592016 expression was positively correlated with BMI, AHI, ODI, TS90; negatively correlated with LSaO2. Among them, this suggests that ENST00000592016 is associated with clinical symptoms of OSA and may serve as a potential molecular target for diagnosis and risk stratification.
The ceRNAs are transcripts that regulate each other at the post-transcriptional level by competing for shared miRNAs. The ceRNA network links the functions of protein-coding mRNAs with those of non-coding RNAs such as miRNAs and lncRNAs (57). Du et al. reported that lncRNA MALAT1 promoted NLRP3 expression by acting as a ceRNA and sponging miR-224-5p, ultimately affecting the NLRP3/IL-1β pathway in the hippocampus. This suggested that lncRNA MALAT1/miR-224-5p/NLRP3 may serve as a potential target for OSA therapy (12). However, to date, there has been no study on ENST00000592016 in OSA. We predicted the binding sites of ENST00000592016-regulated miRNAs interacting with target genes by miRanda. These regulatory axes may provide new targets for the diagnosis and treatment of OSA. However, this requires further in-depth research and verification in the future.
The PI3K/Akt pathway is one of the most important pathways controlling many cellular processes, including cell division, autophagy, survival, and differentiation (58). The MAPK signaling pathway is shared by 4 distinct cascades, including ERK1/2, JNK1/2/3, p38-MAPK, and ERK5. The MAPK/ERK pathway has also been reported to be involved in cell proliferation, differentiation, migration, senescence, and apoptosis (59). Meanwhile, TNF is a cytokine that can directly kill tumor cells and has no obvious cytotoxicity to normal cells. It is a cell signal protein involved in systemic inflammation and one of the cytokines that constitute the acute phase reaction (60). Stimulation or inhibition of the TNF superfamily signaling pathway is expected to yield therapeutic benefits in patients with a variety of diseases, including cancer, autoimmunity, and infectious diseases (61). In an OSA study, genistein protected genioglossus myoblasts from hypoxia-induced injury through the PI3K-Akt and ERK MAPK pathways (62). Ge et al. reported that the lncRNA ROR played a vital role in OSA by regulating miR-145 as well as regulating ERK and MAPK signaling to alleviate CoCl2-induced hypoxic injury (63). Furthermore, Chen et al. showed that low expression of miR-21-5p in OSA patients modulated intermittent hypoxia and reoxygenation-induced apoptosis and cytotoxicity by targeting pro-inflammatory TNF-α-TLR4 signaling (64). These studies suggest an important role for the involved PI3K-Akt, MAPK, and TNF signaling pathways in OSA. However, in the present study, we only predicted that the PI3K-Akt, MAPK, and TNF signaling pathways were involved in the ceRNA regulatory network through miRanda. The mechanism validation of ENST00000592016 involving the PI3K-Akt, MAPK, and TNF signaling pathways still needs to be further elucidated.
Our research provides new targets to explore for the identification of biomarkers for the non-invasive diagnosis of OSA and for developing new treatments. However, there were shortcomings in our current study. The study was performed at a single institution, the number of participants was small; our analysis was diagnostic tests in design, all the patients included in our current study were OSA and HTN-OSA patients before treatment, while the indicators after treatment were not included in the comparison. Therefore, a large, multicenter, prospective, clinical trial would be required to address the above deficiencies.
Conclusions
The current study is the first to study the differential expression of lncRNA in OSA and HTN-OSA exosomes through high-throughput sequencing. A singular lncRNA was identified that specifically differentiates between non-OSA and OSA patients, and the results suggest that ENST00000592016 level is associated with the severity of OSA, which provides preliminary work for the study of potential biomarker of OSA and may help us understand the mechanisms behind OSA development.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3818/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3818/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3818/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This experiment was approved by the Ethics Committee of Nanfang Hospital of Southern Medical University (approval No. NFEC-2021-436) and written informed consent was provided by each patient.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Benjafield AV, Ayas NT, Eastwood PR, et al. Estimation of the global prevalence and burden of obstructive sleep apnoea: a literature-based analysis. Lancet Respir Med 2019;7:687-98. [Crossref] [PubMed]
- Mickelson SA. Oral Appliances for Snoring and Obstructive Sleep Apnea. Otolaryngol Clin North Am 2020;53:397-407. [Crossref] [PubMed]
- Naik V, Khandekar N, Deogaonkar M. Neuromodulation in Obstructive Sleep Apnea. Neurol India 2020;68:S302-6. [Crossref] [PubMed]
- Ucak S, Dissanayake HU, Sutherland K, et al. Heart rate variability and obstructive sleep apnea: Current perspectives and novel technologies. J Sleep Res 2021;30:e13274. [Crossref] [PubMed]
- Diamond JA, Ismail H. Obstructive Sleep Apnea and Cardiovascular Disease. Clin Geriatr Med 2021;37:445-56. [Crossref] [PubMed]
- Patel AR, Patel AR, Singh S, et al. The Association of Obstructive Sleep Apnea and Hypertension. Cureus 2019;11:e4858. [Crossref] [PubMed]
- Martínez-García MA, Capote F, Campos-Rodríguez F, et al. Effect of CPAP on blood pressure in patients with obstructive sleep apnea and resistant hypertension: the HIPARCO randomized clinical trial. JAMA 2013;310:2407-15. [Crossref] [PubMed]
- Khurshid K, Yabes J, Weiss PM, et al. Effect of Antihypertensive Medications on the Severity of Obstructive Sleep Apnea: A Systematic Review and Meta-Analysis. J Clin Sleep Med 2016;12:1143-51. [Crossref] [PubMed]
- Puvvula PK. LncRNAs Regulatory Networks in Cellular Senescence. Int J Mol Sci 2019;20:2615. [Crossref] [PubMed]
- Chen Q, Lin G, Huang J, et al. Expression profile of long non-coding RNAs in rat models of OSA-induced cardiovascular disease: new insight into pathogenesis. Sleep Breath 2019;23:795-804. [Crossref] [PubMed]
- Zhang Z, Li Z, Wang Y, et al. Overexpressed long noncoding RNA CPS1-IT alleviates pulmonary arterial hypertension in obstructive sleep apnea by reducing interleukin-1β expression via HIF1 transcriptional activity. J Cell Physiol 2019;234:19715-27. [Crossref] [PubMed]
- Du P, Wang J, Han Y, et al. Blocking the LncRNA MALAT1/miR-224-5p/NLRP3 Axis Inhibits the Hippocampal Inflammatory Response in T2DM With OSA. Front Cell Neurosci 2020;14:97. [Crossref] [PubMed]
- Khalyfa A, Kheirandish-Gozal L, Gozal D. Circulating exosomes in obstructive sleep apnea as phenotypic biomarkers and mechanistic messengers of end-organ morbidity. Respir Physiol Neurobiol 2018;256:143-56. [Crossref] [PubMed]
- Lin J, Li J, Huang B, et al. Exosomes: novel biomarkers for clinical diagnosis. ScientificWorldJournal 2015;2015:657086. [Crossref] [PubMed]
- Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science 2020;367:eaau6977. [Crossref] [PubMed]
- Khalyfa A, Marin JM, Qiao Z, et al. Plasma exosomes in OSA patients promote endothelial senescence: effect of long-term adherent continuous positive airway pressure. Sleep 2020;43:zsz217. [Crossref] [PubMed]
- Teng Y, Kang H, Chu Y. Identification of an Exosomal Long Noncoding RNA SOX2-OT in Plasma as a Promising Biomarker for Lung Squamous Cell Carcinoma. Genet Test Mol Biomarkers 2019;23:235-40. [Crossref] [PubMed]
- Zheng P, Zhang H, Gao H, et al. Plasma Exosomal Long Noncoding RNA lnc-SLC2A12-10:1 as a Novel Diagnostic Biomarker for Gastric Cancer. Onco Targets Ther 2020;13:4009-18. [Crossref] [PubMed]
- Hu X, Wu D, He X, et al. circGSK3β promotes metastasis in esophageal squamous cell carcinoma by augmenting β-catenin signaling. Mol Cancer 2019;18:160. [Crossref] [PubMed]
- Du Y, Wang X, Li L, et al. miRNA-Mediated Suppression of a Cardioprotective Cardiokine as a Novel Mechanism Exacerbating Post-MI Remodeling by Sleep Breathing Disorders. Circ Res 2020;126:212-28. [Crossref] [PubMed]
- Zhu L, Chamberlin NL, Arrigoni E. Muscarinic Inhibition of Hypoglossal Motoneurons: Possible Implications for Upper Airway Muscle Hypotonia during REM Sleep. J Neurosci 2019;39:7910-9. [Crossref] [PubMed]
- Kyotani Y, Ota H, Itaya-Hironaka A, et al. Intermittent hypoxia induces the proliferation of rat vascular smooth muscle cell with the increases in epidermal growth factor family and erbB2 receptor. Exp Cell Res 2013;319:3042-50. [Crossref] [PubMed]
- Zhang CQ, Yi S, Chen BB, et al. mTOR/NF-κB signaling pathway protects hippocampal neurons from injury induced by intermittent hypoxia in rats. Int J Neurosci 2021;131:994-1003. [Crossref] [PubMed]
- Pan YY, Deng Y, Xie S, et al. Altered Wnt Signaling Pathway in Cognitive Impairment Caused by Chronic Intermittent Hypoxia: Focus on Glycogen Synthase Kinase-3β and β-catenin. Chin Med J (Engl) 2016;129:838-45. [Crossref] [PubMed]
- Calhoun DA. Obstructive sleep apnea and hypertension. Curr Hypertens Rep 2010;12:189-95. [Crossref] [PubMed]
- Das AM, Khayat R. Hypertension in obstructive sleep apnea: risk and therapy. Expert Rev Cardiovasc Ther 2009;7:619-26. [Crossref] [PubMed]
- Huang X, Yuan T, Tschannen M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics 2013;14:319. [Crossref] [PubMed]
- Wang J, Liu Y, Sun W, et al. Plasma exosomes as novel biomarker for the early diagnosis of gastric cancer. Cancer Biomark 2018;21:805-12. [Crossref] [PubMed]
- Hu D, Zhan Y, Zhu K, et al. Plasma Exosomal Long Non-Coding RNAs Serve as Biomarkers for Early Detection of Colorectal Cancer. Cell Physiol Biochem 2018;51:2704-15. [Crossref] [PubMed]
- Wang D, Wang P, Bian X, et al. Elevated plasma levels of exosomal BACE1-AS combined with the volume and thickness of the right entorhinal cortex may serve as a biomarker for the detection of Alzheimer's disease. Mol Med Rep 2020;22:227-38. [Crossref] [PubMed]
- Karp PD, Midford PE, Caspi R, et al. Pathway size matters: the influence of pathway granularity on over-representation (enrichment analysis) statistics. BMC Genomics 2021;22:191. [Crossref] [PubMed]
- Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353-61. [Crossref] [PubMed]
- Kanehisa M, Sato Y, Kawashima M, et al. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 2016;44:D457-62. [Crossref] [PubMed]
- Vander Ark A, Cao J, Li X. TGF-β receptors: In and beyond TGF-β signaling. Cell Signal 2018;52:112-20. [Crossref] [PubMed]
- Cubillos-Zapata C, Martínez-García MÁ, Díaz-García E, et al. Obesity attenuates the effect of sleep apnea on active TGF-ß1 levels and tumor aggressiveness in patients with melanoma. Sci Rep 2020;10:15528. [Crossref] [PubMed]
- Ljunggren M, Theorell-Haglöw J, Freyhult E, et al. Association between proteomics and obstructive sleep apnea phenotypes in a community-based cohort of women. J Sleep Res 2020;29:e13041. [Crossref] [PubMed]
- Iovino L, Tremblay ME, Civiero L. Glutamate-induced excitotoxicity in Parkinson's disease: The role of glial cells. J Pharmacol Sci 2020;144:151-64. [Crossref] [PubMed]
- He S, Zhang X, Qu S. Glutamate, Glutamate Transporters, and Circadian Rhythm Sleep Disorders in Neurodegenerative Diseases. ACS Chem Neurosci 2019;10:175-81. [Crossref] [PubMed]
- Yu X, Li W, Ma Y, et al. GABA and glutamate neurons in the VTA regulate sleep and wakefulness. Nat Neurosci 2019;22:106-19. [Crossref] [PubMed]
- He Y, Xiang L, Zhao LP, et al. Relationship between soluble Semaphorin4D and cognitive impairment in patients with obstructive sleep apnea-hypopnea syndrome. Eur Arch Otorhinolaryngol 2017;274:1263-8. [Crossref] [PubMed]
- Ralls F, Cutchen L. A contemporary review of obstructive sleep apnea. Curr Opin Pulm Med 2019;25:578-93. [Crossref] [PubMed]
- Kheirandish-Gozal L, McManus CJT, Kellermann GH, et al. Urinary neurotransmitters are selectively altered in children with obstructive sleep apnea and predict cognitive morbidity. Chest 2013;143:1576-83. [Crossref] [PubMed]
- Ledonne A, Mercuri NB. On the Modulatory Roles of Neuregulins/ErbB Signaling on Synaptic Plasticity. Int J Mol Sci 2019;21:275. [Crossref] [PubMed]
- Zhu J, Zhu Z, Ren Y, et al. Role of the Nrdp1 in Brain Injury Induced by Chronic Intermittent Hypoxia in Rats via Regulating the Protein Levels of ErbB3. Neurotox Res 2020;38:124-32. [Crossref] [PubMed]
- Li Y, Zhang J, Pan J, et al. Integrated bioinformatical analysis of lncRNA-mRNA co-expression profiles of cervical cancer. CEOG 2021;48:1381-92. [Crossref]
- Mei ZG, Huang YG, Feng ZT, et al. Electroacupuncture ameliorates cerebral ischemia/reperfusion injury by suppressing autophagy via the SIRT1-FOXO1 signaling pathway. Aging (Albany NY) 2020;12:13187-205. [Crossref] [PubMed]
- Zhang X, Wei M, Fan J, et al. Ischemia-induced upregulation of autophagy preludes dysfunctional lysosomal storage and associated synaptic impairments in neurons. Autophagy 2021;17:1519-42. [Crossref] [PubMed]
- Mossmann D, Park S, Hall MN. mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat Rev Cancer 2018;18:744-57. [Crossref] [PubMed]
- Switon K, Kotulska K, Janusz-Kaminska A, et al. Molecular neurobiology of mTOR. Neuroscience 2017;341:112-53. [Crossref] [PubMed]
- Ko CY, Su HZ, Zhang L, et al. Disturbances of the Gut Microbiota, Sleep Architecture, and mTOR Signaling Pathway in Patients with Severe Obstructive Sleep Apnea-Associated Hypertension. Int J Hypertens 2021;2021:9877053. [Crossref] [PubMed]
- Zhou S, Xia L, Han L. SFRP1 suppresses granulosa cell proliferation and migration through inhibiting JNK pathway. CEOG 2021;48:1193-9.
- Li H, Bai R, Zhao Z, et al. Application of droplet digital PCR to detect the pathogens of infectious diseases. Biosci Rep 2018;38:BSR20181170. [Crossref] [PubMed]
- Nyaruaba R, Mwaliko C, Kering KK, et al. Droplet digital PCR applications in the tuberculosis world. Tuberculosis (Edinb) 2019;117:85-92. [Crossref] [PubMed]
- Chiu HY, Chen PY, Chuang LP, et al. Diagnostic accuracy of the Berlin questionnaire, STOP-BANG, STOP, and Epworth sleepiness scale in detecting obstructive sleep apnea: A bivariate meta-analysis. Sleep Med Rev 2017;36:57-70. [Crossref] [PubMed]
- Deng M, Huang YT, Xu JQ, et al. Association Between Intermittent Hypoxia and Left Ventricular Remodeling in Patients With Obstructive Sleep Apnea-Hypopnea Syndrome. Front Physiol 2021;11:608347. [Crossref] [PubMed]
- Lisi E, Faini A, Bilo G, et al. Diastolic dysfunction in controlled hypertensive patients with mild-moderate obstructive sleep apnea. Int J Cardiol 2015;187:686-92. [Crossref] [PubMed]
- Qi X, Zhang DH, Wu N, et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet 2015;52:710-8. [Crossref] [PubMed]
- Jafari M, Ghadami E, Dadkhah T, et al. PI3k/AKT signaling pathway: Erythropoiesis and beyond. J Cell Physiol 2019;234:2373-85. [Crossref] [PubMed]
- Sun Y, Liu WZ, Liu T, et al. Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res 2015;35:600-4. [Crossref] [PubMed]
- Monaco C, Nanchahal J, Taylor P, et al. Anti-TNF therapy: past, present and future. Int Immunol 2015;27:55-62. [Crossref] [PubMed]
- Vanamee ÉS, Faustman DL. Structural principles of tumor necrosis factor superfamily signaling. Sci Signal 2018;11:aao4910. [Crossref] [PubMed]
- Ding W, Chen X, Li W, et al. Genistein Protects Genioglossus Myoblast Against Hypoxia-induced Injury through PI3K-Akt and ERK MAPK Pathways. Sci Rep 2017;7:5085. [Crossref] [PubMed]
- Ge H, Liu J, Liu F, et al. Long non-coding RNA ROR mitigates cobalt chloride-induced hypoxia injury through regulation of miR-145. Artif Cells Nanomed Biotechnol 2019;47:2221-9. [Crossref] [PubMed]
- Chen YC, Hsu PY, Su MC, et al. miR-21-5p Under-Expression in Patients with Obstructive Sleep Apnea Modulates Intermittent Hypoxia with Re-Oxygenation-Induced-Cell Apoptosis and Cytotoxicity by Targeting Pro-Inflammatory TNF-α-TLR4 Signaling. Int J Mol Sci 2020;21:999. [Crossref] [PubMed]
(English Language Editor: J. Jones)


