
Exploration of immune infiltration and feature genes in viral hepatitis-associated liver fibrosis using transcriptome data
Introduction
Liver fibrosis is caused by continuous and repeated liver damage after viral infection. Progressive liver fibrosis without effective prevention and treatment can progress to cirrhosis and liver cancer, which ranks fifth in incidence among cancer cases globally (1). With early diagnosis and treatment, the progression of liver fibrosis can be prevented or reversed (2,3). Therefore, early detection, dynamic assessment, and effective intervention to prevent continued damage to the liver are crucial for improving the prognosis of patients with chronic liver disease.
Innate and adaptive immune alterations resulting from viral infections are vital to the pathogenesis of hepatic fibrosis (4,5). Hepatic stellate cells (HSC) are precursors of myofibroblasts, which play an important role in liver fibrosis (6). Immune cells participate in connective tissue proliferation, contributing to liver fibrosis. However, the immune mechanisms leading to viral liver fibrosis need to be further explored. In addition, it is crucial to discover differential genes and explore possible related mechanisms in viral hepatitis-associated liver fibrosis.
In this study, we aimed to systematically explore immune cell infiltration using the Gene Expression Omnibus (GEO) database predicted by CIBERSORT and to identify potential feature genes, thereby providing new insights into the treatment of viral hepatitis-associated liver fibrosis.
Methods
Databases
Gene expression profiles were selected from the GEO database (Entrez GEO Profiles, RRID: SCR_004584, archived on March 21, 2022). The inclusion criteria were human liver tissue containing hepatitis B or C virus-related liver fibrosis and non-fibrosis, and expression profiling by microarray. Finally, we included the datasets GSE14323, GSE33650, and GSE6764 for testing, and GSE84044 with the largest sample size for validation. Detailed information is shown in Table 1. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Table 1
| Group | GEO accession | Platform | Tissue (Homo sapiens) | Sample number | ||
|---|---|---|---|---|---|---|
| Total | Fibrosis | Without fibrosis | ||||
| Testing | GSE14323 | GPL571 | Liver | 60 | 41 | 19 |
| Testing | GSE33650 | GPL14877 | Liver | 9 | 4 | 5 |
| Testing | GSE6764 | GPL570 | Liver | 23 | 13 | 10 |
| Validation | GSE84044 | GPL570 | Liver | 124 | 81 | 43 |
GEO, Gene Expression Omnibus.
Data merging and batch correction
R software (v. 4.1.3, R Project for Statistical Computing, RRID: SCR_001905) was utilized for data analysis and visualization. After merging GSE14323, GSE33650, and GSE6764 gene matrices using the Perl script, the R package “sva” was applied to remove the heterogeneity caused by different experimental batches and platforms. Corrected gene expression matrices were analyzed using the R package “limma”. For the merged probe matrix files, each probe expression matrix was extracted using Perl scripts and transformed into gene expression matrices using the platform files. To verify the correction of the batch effect, principal component analysis was performed on the merged probe matrices before and after merging. The results of the sample clustering analysis were displayed visually using the R package “ggplot2”.
Assessment of immune cell infiltration
Using the collated gene expression matrix, immune cell composition was calculated using the CIBERSORT (CIBERSORT, RRID: SCR_016955, archived on March 24, 2022) algorithm to determine immune infiltration. The gene matrix was re-corrected and the p-value for each sample for immune cell determination was detected using CIBERSORT. Only data with P values <0.05 were retained for subsequent analysis. To perform calculations, the characteristic gene set of LM22 provided by CIBERSORT was used as a reference for the immune cell subgroups. CIBERSORT results were visually presented using the R package “corrplot”, “vioplot”.
Identification of characteristic immune cell subgroups by least absolute shrinkage and selection operator (LASSO) regression
Data obtained using the CIBERSORT algorithm were further analyzed. LASSO regression was used to identify immune cell subgroups that were distinguished from liver fibrosis and non-fibrosis. Immune cell subgroups with P values <0.05, determined by Wilcoxon test, were intersected with the characteristic immune cell subgroups obtained from LASSO regression to obtain the most accurate characteristic subgroups possible.
Differentially expressed genes (DEGs) analysis
We used the “remove batch effect” function in the R package “limma” and the R package “sva” for batch correction to find DEGs. Both methods selected genes with log FC >1 and adj.P.Val <0.05 as differential genes. Finally, the obtained DEGs were considered as intersection sets and represented using Venn diagrams. The heatmap and Venn diagram for results visualization were plotted using the R package “heatmap” with “VennDiagram”.
DEGs enrichment analysis
The gene names of DEGs were converted to gene IDs using the R package “org.Hs.eg.db” and analyzed for Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment. GO and KEGG enrichment analyses were performed using the R package “clusterProfiler”. Pathways were considered significant at P values <0.05 and q values <0.05. The results were visualized using the R packages, “enrichplot” and “ggplot2”.
Hub genes search
STRING (STRING, RRID: SCR_005223, archived on March 28, 2022) was used for protein-protein interaction (PPI) network analysis. Only those interactions with a score >0.7 were included. The number of PPI nodes was sorted, and the top five genes were considered hub genes. The results of the top 30 are represented in a bar graph.
Receiver operator characteristics (ROC) analysis
ROC analysis was performed to determine the predictive value of hub genes with dataset GSE84044 using IBM SPSS Statistics (version 25, IBM SPSS Statistics, RRID: SCR_016479).
Analysis of the correlation between hub genes and characteristic immune cell subgroups
Immune infiltration analysis was performed for GSE84044 using the CIBERSORT algorithm in the same manner as described above. Spearman correlation analysis in R was used to explore the correlation between hub genes and levels of infiltrative immune cells. Correlations were visualized using the R package “ggplot2”.
Results
Immune cell infiltration in fibrosis and non-fibrosis liver tissues
We selected GSE14323, GSE33650, and GSE6764 for analysis, which included 58 liver fibrotic tissues and 34 non-fibrotic tissues. Data were corrected using Principal Component Analysis (PCA) (Figure S1). Finally, all data conformed to CIBERSORT analysis (P<0.05). The 22 immune cell categories included CD8+ T cells, naive and memory resting and memory activated CD4+ T cells, Treg cells, Tfh cells, gamma-delta T cells, M0–2 macrophages, resting and activated natural killer (NK) cells, resting and activated mast cells, naive and memory B cells, resting and activated dendritic cells, neutrophils, monocytes, plasma cells, and eosinophils. The results are shown in histograms and heat maps (Figure 1A-1C). The results showed that M0–2 macrophages, naive B cells, CD8+ T cells, resting CD4+ memory T cells, Tfh cells, gamma-delta T cells, activated NK cells, resting mast cells, and plasma cells were the main infiltrative immune cells represented.
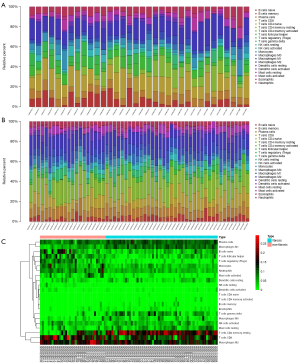
The correlation between 22 infiltrative immune cells in fibrotic and non-fibrotic tissues is shown in Figure 2. In non-fibrotic tissues, resting CD4+ memory T cells showed a significantly positive correlation with M1 macrophages, but showed a negative correlation in fibrotic tissues. Neutrophils were positively correlated with M0 macrophages in non-fibrotic tissues but this correlation was weakened in fibrotic tissues while resting and activated mast cells had a significantly negative correlation in both fibrotic and non-fibrotic tissues.
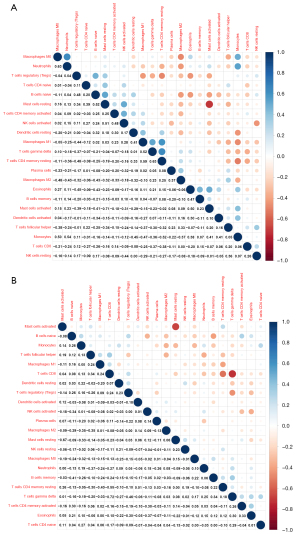
We used two methods to identify different infiltrations of immune cells in fibrosis and non-fibrosis. The Wilcoxon test showed 17 immune cells (P<0.05) in Figure 3A, and the LASSO text showed 11 cell types (Figure 3B,3C). The intersection of the two methods yielded 10 types of immune cells: naive B cells, plasma cells, resting CD4+ memory T cells, Tfh cells, Treg cells, M0-M2 macrophages, and resting and activated mast cells.
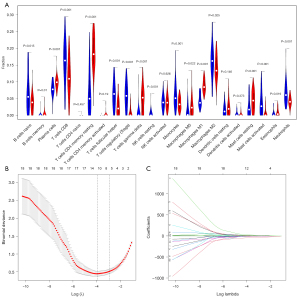
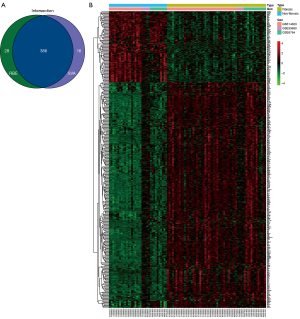
Identification of DEGs and functional enrichment analysis
The PPI network generated using STRING is shown in Figure S2. A total of 58 liver fibrotic tissues and 34 non-fibrotic tissues were used to identify DEGs. We obtained 98 upregulated and 304 downregulated genes using the “remove batch effect”, whereas 106 upregulated and 308 downregulated genes were obtained using the R package “sva”. The intersection of the two methods revealed 386 DEGs, including 92 upregulated and 294 downregulated genes. The results are shown in Figure 4A,4B.
GO was used to explore the functional enrichment of DEGs (Figure 5A,5B). DEGs were mainly associated with antiviral responses. KEGG analysis was performed to analyze the relationship between DEGs and signaling pathways (Figure 5C,5D). DEGs were mainly related to viral protein-cytokines interaction, indicating that they were closely associated with antiviral responses.
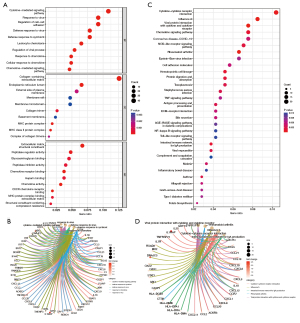
Identification and validation of hub genes
Three datasets were used to identify hub genes. The top five genes were selected. As three genes were ranked 4th in PPI, the six genes with the top four PPI numbers were selected. We identified the first six significant hub genes, including STAT1, CXCL10, PTPRC, IFIT3, OAS2, and MX1 (Figure S3). All six genes were upregulated. The expression levels of the six hub genes in the three test datasets are shown in Figure 6A. Expression verification based on GSE84044 is presented in the heatmap (Figure 6B).
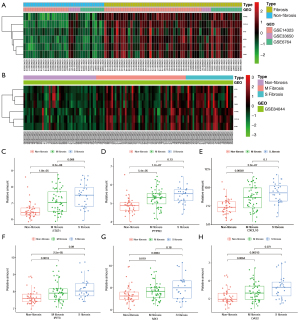
The samples were divided into three groups based on their Scheuer scores. S0 of the Scheuer score was defined as non-fibrosis, S1–2 as mild to moderate fibrosis (M fibrosis), and S3–4 as severe fibrosis (S fibrosis). STAT1, CXCL10, PTPRC, IFIT3, OAS2, and MX1 were used to identify liver fibrosis, while STAT1, CXCL10, and PTPRC were used to identify early liver fibrosis. The expression of the six hub genes in different tissues is shown in Figure 6C-6H.
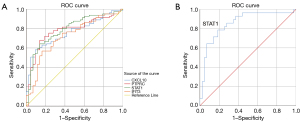
Diagnostic effectiveness of hub genes
ROC analysis was used to verify the diagnostic validity of the hub genes for liver fibrosis and non-fibrosis (Figure 7). Considering an area under the curve (AUC) >0.7 (P<0.05) of diagnostic value, STAT1 (AUC =0.790, P=0.42, 95% CI: 0.707–0.873), CXCL10 (AUC =0.760, P=0.43, 95% CI: 0.676–0.844), PTPRC (AUC =0.778, P=0.44, 95% CI: 0.692–0.863) and IFIT3 (AUC =0.721, P=0.48, 95% CI: 0.627–0.815) had adequate sensitivity and specificity. In addition, STAT1 had a strong diagnostic ability for non-fibrosis and severe fibrosis with an AUC of 0.855 (P=0.048, 95% CI: 0.761–0.950).
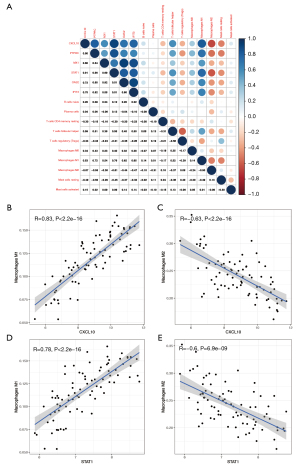
Correlation between hub genes and differential infiltrating immune cells
GSE84044 was used to identify correlations between hub genes and differential infiltrating immune cells predicted by CIBERSORT (Figure 8A). Surprisingly, all six hub genes were correlated with M1 and M2 macrophages. The correlation with M1 macrophages was stronger than that with M2 macrophages. Moreover, six hub genes were positively correlated with M1 macrophages and negatively correlated with M2 macrophages. Detailed information is shown in Figure 8B-8E and Figure S4A-S4H.
Discussion
In this study, we systematically investigated the infiltration of immune cells into viral hepatitis-associated liver fibrosis and explored the role of immune cells in the progression of fibrosis. In addition, we identified six hub genes and identified their association with infiltrating immune cells.
In fibrotic liver tissues, plasma cells, resting CD4+ memory T cells, M0–1 macrophages, and resting mast cells showed higher counts; meanwhile, naive B cells, Tfh, Tregs, M2 macrophages, and activated mast cells showed lower counts than those in non-fibrotic liver tissues. M0 macrophages can polarize to M1 and M2, and the balanced relationship between M1 and M2 is closely related to inflammatory damage and tissue repair. M1 may mediate pathogen clearance and exacerbate tissue damage, while M2 inhibits the inflammatory response and promotes tissue repair and remodeling (7). In the current literature, it remains controversial whether macrophages in M1 or M2 promote or inhibit fibrosis (8-12). In our study, M1 levels were higher and M2 macrophage levels were lower in the fibrotic tissues. Upon hepatitis virus activity, the organism reactively secretes molecules such as interferon (IFN)-γ and tumor necrosis factor (TNF), which contribute to the polarization of M0 into M1 macrophages. Polarized M1 macrophages have powerful phagocytic and antigen-presenting functions but also release large amounts of inflammatory factors such as TNF and interleukin (IL)-6 (13). Due to the chronic activity of the hepatitis virus, M1 macrophages remain high for a long time. The persistence of high levels of M1 macrophages also enables chronic inflammation in the liver. Its prolonged existence prevents inflammation from subsiding, causes scarring, and promotes fibrosis (14). M2 macrophages balance and regulate the inflammation produced by M1 macrophages. The effect of this balance on the outcome of tissues and organs has been elucidated in pulmonary fibrosis, renal fibrosis, and atherosclerosis (15-17). We hypothesize that the imbalance between persistent M1 macrophages and relatively deficient M2 macrophages due to chronic infection has led to the progressive development of fibrosis in patients with viral hepatitis. After antigen presentation by M1 macrophages, naive B cells are activated as plasma cells, which play a vital role in virus clearance (18). Interestingly, we found that plasma cell levels were significantly elevated in fibrotic tissues. B cells are known to promote and maintain liver fibrosis by regulating inflammation and limiting HSC senescence (19). CD4+ T cells have several subpopulations, including Th1, Th2, Th17, Tregs, and Tfh cells. They also perform various supplementary immune functions (20). Our results showed that the total CD4+ cell count was elevated in fibrotic tissues, but Treg and Tfh cell levels were lower than those in non-fibrotic tissues. Tregs exert anti-inflammatory effects and regulate immune homeostasis. They interact with Th17 cells. Previous studies have shown that Tregs control liver fibrosis progression, promote tissue repair, and restore tissue integrity (21,22). Tfh cells exert their functions through auxiliary B cells (23). It has been suggested that Tfh cells may have granuloma-inducing and pro-liver fibrosis effects (24). In summary, during the persistence of viral hepatitis, inflammation-associated immune cells were upregulated, and anti-inflammatory immune cells were downregulated. The imbalance in immune cells allows liver fibrosis to occur and persist.
Our study identified STAT1, CXCL10, PTPRC, IFIT3, OAS2, and MX1 as hub genes in viral hepatitis-associated liver fibrosis. All these genes were upregulated in fibrosis and were closely associated with the progression of fibrosis.
STAT1 mediates IFN and IL-27 signaling, and regulates cell proliferation, differentiation, and survival. It is an integral part of the protective immunity against pathogens (25). It has been shown that STAT1 may reduce liver fibrosis by inhibiting HSC proliferation and stimulating NK cells to destroy activated HSC (26). CXCL10 can have an expressional induction after viral stimulation of the liver (27). Several studies have suggested that CXCL10 is associated with viral liver fibrosis (28,29). A study revealed that CXCL10 interferes with NK cell-mediated inactivation of HSC, which in turn promotes liver fibrosis (30). It has also been shown that CXCL10 induces the hepatic expression of IL-9 and activates the Raf/MEK/ERK signaling pathway to promote liver fibrosis (31). PTPRC is an essential component in the regulation of innate immune signaling pathways and plays a role in signaling to T- and B-cell receptors (32). It has been suggested that PTPRC is an effective marker for the identification of fibrosis (33). IFIT3 is an interferon-inducible protein with a tetrapeptide repeat sequence. Various pathogens, especially DNA and RNA viruses, can induce IFIT3 expression, and IFIT3 performs antiviral defense (34,35). However, the mechanism of its interaction with viral hepatitis-associated liver fibrosis needs to be further investigated. Additionally, OAS2 and MX1 play important roles in the progression of liver fibrosis (36,37). Overall, these genes may play important roles in viral hepatitis-associated liver fibrosis and contribute to fibrosis development.
Finally, we analyzed the correlation between immune cells and hub genes. Interestingly, all six genes were closely related to M1 and M2 macrophages. Moreover, these hub genes were surprisingly consistent in their correlation with M1 and M2 macrophages, as they were positively correlated with M1 macrophages and negatively correlated with M2 macrophages. It has been found that IFN-mediated STAT1 induces macrophage polarization toward M1 (38). A previous study indicated that M1 macrophages secrete pro-inflammatory interleukin 6 and induce CXCL10 expression, thereby promoting inflammation (39). Another study revealed that CXCL10 enhances the expression of M1 macrophage phenotype markers and decreases the expression of M2 markers, suggesting that CXCL10 is prone to induce macrophage polarization to M1 (40). It is very likely that the six hub genes stimulate M0 macrophages to M1 polarization and suppress M2 levels through various pathways, thus promoting fibrosis formation. This suggests that macrophages play a crucial role in the immune process of fibrosis.
Nevertheless, this study has some limitations. First, only bioinformatic analyses were performed because of sample limitations. Immune cell analysis was based on the CIBERSORT algorithm. Although many studies have demonstrated its relative accuracy, further experiments, such as flow cytometric sorting, are needed to confirm the immune cell infiltration in the liver tissue. In addition, owing to the lack of clinical information and the sample size limitation, no further classification studies were conducted with patient basic information or their treatment methods.
Conclusions
In this study, we systematically analyzed immune cell infiltration in viral hepatitis-associated liver fibrosis tissues and identified six hub genes. These genes are closely associated with the balance between M1 and M2 macrophages, which provides a new perspective on the mechanisms of liver fibrosis.
Acknowledgments
Funding: This work was supported by the 13th Five-Year Plan of the Ministry of Science and Technology of the People’s Republic of China (No. 2017ZX10203202–003) and Beijing Municipal Science and Technology Commission of Major Projects (No. D161100002716003).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-2205/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Campana L, Iredale JP. Regression of Liver Fibrosis. Semin Liver Dis 2017;37:1-10. [Crossref] [PubMed]
- Xu XY, Ding HG, Li WG, et al. Chinese guidelines on the management of liver cirrhosis (abbreviated version). World J Gastroenterol 2020;26:7088-103. [Crossref] [PubMed]
- Zaki MYW, Fathi AM, Samir S, et al. Innate and Adaptive Immunopathogeneses in Viral Hepatitis; Crucial Determinants of Hepatocellular Carcinoma. Cancers (Basel) 2022;14:1255. [Crossref] [PubMed]
- Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV- and HCV-associated hepatocellular carcinoma. Nat Rev Cancer 2013;13:123-35. [Crossref] [PubMed]
- Higashi T, Friedman SL, Hoshida Y. Hepatic stellate cells as key target in liver fibrosis. Adv Drug Deliv Rev 2017;121:27-42. [Crossref] [PubMed]
- Wang C, Ma C, Gong L, et al. Macrophage Polarization and Its Role in Liver Disease. Front Immunol 2021;12:803037. [Crossref] [PubMed]
- Rao J, Wang H, Ni M, et al. FSTL1 promotes liver fibrosis by reprogramming macrophage function through modulating the intracellular function of PKM2. Gut 2022; Epub ahead of print. [Crossref] [PubMed]
- Cui W, Zhou S, Wang Y, et al. Cadmium exposure activates the PI3K/AKT signaling pathway through miRNA-21, induces an increase in M1 polarization of macrophages, and leads to fibrosis of pig liver tissue. Ecotoxicol Environ Saf 2021; Epub ahead of print. [Crossref] [PubMed]
- Liu P, Li H, Gong J, et al. Chitooligosaccharides alleviate hepatic fibrosis by regulating the polarization of M1 and M2 macrophages. Food Funct 2022;13:753-68. [Crossref] [PubMed]
- Beljaars L, Schippers M, Reker-Smit C, et al. Hepatic Localization of Macrophage Phenotypes during Fibrogenesis and Resolution of Fibrosis in Mice and Humans. Front Immunol 2014;5:430. [Crossref] [PubMed]
- Xi S, Zheng X, Li X, et al. Activated Hepatic Stellate Cells Induce Infiltration and Formation of CD163+ Macrophages via CCL2/CCR2 Pathway. Front Med (Lausanne) 2021;8:627927. [Crossref] [PubMed]
- Wang S, Liu R, Yu Q, et al. Metabolic reprogramming of macrophages during infections and cancer. Cancer Lett 2019;452:14-22. [Crossref] [PubMed]
- Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature 2013;496:445-55. [Crossref] [PubMed]
- Chang X, Xing L, Wang Y, et al. Nanoengineered immunosuppressive therapeutics modulating M1/M2 macrophages into the balanced status for enhanced idiopathic pulmonary fibrosis therapy. Nanoscale 2020;12:8664-78. [Crossref] [PubMed]
- Xie Y, Hu X, Li S, et al. Pharmacological targeting macrophage phenotype via gut-kidney axis ameliorates renal fibrosis in mice. Pharmacol Res 2022;178:106161. [Crossref] [PubMed]
- Li Z, Ma D, Wang Y, et al. Astragali Radix-Coptis Rhizoma Herb Pair Attenuates Atherosclerosis in ApoE-/- Mice by Regulating the M1/M2 and Th1/Th2 Immune Balance and Activating the STAT6 Signaling Pathway. Evid Based Complement Alternat Med 2022;2022:7421265. [Crossref] [PubMed]
- Khanam A, Chua JV, Kottilil S. Immunopathology of Chronic Hepatitis B Infection: Role of Innate and Adaptive Immune Response in Disease Progression. Int J Mol Sci 2021;22:5497. [Crossref] [PubMed]
- Faggioli F, Palagano E, Di Tommaso L, et al. B lymphocytes limit senescence-driven fibrosis resolution and favor hepatocarcinogenesis in mouse liver injury. Hepatology 2018;67:1970-85. [Crossref] [PubMed]
- Jeong J, Choi YJ, Lee HK. The Role of Autophagy in the Function of CD4+ T Cells and the Development of Chronic Inflammatory Diseases. Front Pharmacol 2022;13:860146. [Crossref] [PubMed]
- Ikeno Y, Ohara D, Takeuchi Y, et al. Foxp3+ Regulatory T Cells Inhibit CCl4-Induced Liver Inflammation and Fibrosis by Regulating Tissue Cellular Immunity. Front Immunol 2020;11:584048. [Crossref] [PubMed]
- Hann A, Oo YH, Perera MTPR. Regulatory T-Cell Therapy in Liver Transplantation and Chronic Liver Disease. Front Immunol 2021;12:719954. [Crossref] [PubMed]
- Walker LSK. The link between circulating follicular helper T cells and autoimmunity. Nat Rev Immunol 2022; Epub ahead of print. [Crossref] [PubMed]
- Beurier P, Ricard L, Eshagh D, et al. TFH cells in systemic sclerosis. J Transl Med 2021;19:375. [Crossref] [PubMed]
- Boisson-Dupuis S, Kong XF, Okada S, et al. Inborn errors of human STAT1: allelic heterogeneity governs the diversity of immunological and infectious phenotypes. Curr Opin Immunol 2012;24:364-78. [Crossref] [PubMed]
- Jeong WI, Park O, Radaeva S, et al. STAT1 inhibits liver fibrosis in mice by inhibiting stellate cell proliferation and stimulating NK cell cytotoxicity. Hepatology 2006;44:1441-51. [Crossref] [PubMed]
- Song X, Gao X, Wang Y, et al. HCV Core Protein Induces Chemokine CCL2 and CXCL10 Expression Through NF-κB Signaling Pathway in Macrophages. Front Immunol 2021;12:654998. [Crossref] [PubMed]
- Singh KP, Zerbato JM, Zhao W, et al. Intrahepatic CXCL10 is strongly associated with liver fibrosis in HIV-Hepatitis B co-infection. PLoS Pathog 2020;16:e1008744. [Crossref] [PubMed]
- Zhang J, Liu Y, Chen H, et al. MyD88 in hepatic stellate cells enhances liver fibrosis via promoting macrophage M1 polarization. Cell Death Dis 2022;13:411. [Crossref] [PubMed]
- Hintermann E, Bayer M, Pfeilschifter JM, et al. CXCL10 promotes liver fibrosis by prevention of NK cell mediated hepatic stellate cell inactivation. J Autoimmun 2010;35:424-35. [Crossref] [PubMed]
- Guo X, Cen Y, Wang J, et al. CXCL10-induced IL-9 promotes liver fibrosis via Raf/MEK/ERK signaling pathway. Biomed Pharmacother 2018;105:282-9. [Crossref] [PubMed]
- Al Barashdi MA, Ali A, McMullin MF, et al. Protein tyrosine phosphatase receptor type C (PTPRC or CD45). J Clin Pathol 2021;74:548-52. [Crossref] [PubMed]
- de Oliveira RC, Wilson SE. Fibrocytes, Wound Healing, and Corneal Fibrosis. Invest Ophthalmol Vis Sci 2020;61:28. [Crossref] [PubMed]
- Chikhalya A, Dittmann M, Zheng Y, et al. Human IFIT3 Protein Induces Interferon Signaling and Inhibits Adenovirus Immediate Early Gene Expression. mBio 2021;12:e0282921. [Crossref] [PubMed]
- Xu F, Song H, An B, et al. NF-κB-Dependent IFIT3 Induction by HBx Promotes Hepatitis B Virus Replication. Front Microbiol 2019;10:2382. [Crossref] [PubMed]
- Leisching G, Wiid I, Baker B. The Association of OASL and Type I Interferons in the Pathogenesis and Survival of Intracellular Replicating Bacterial Species. Front Cell Infect Microbiol 2017;7:196. [Crossref] [PubMed]
- García-Álvarez M, Berenguer J, Jiménez-Sousa MA, et al. Mx1, OAS1 and OAS2 polymorphisms are associated with the severity of liver disease in HIV/HCV-coinfected patients: A cross-sectional study. Sci Rep 2017;7:41516. [Crossref] [PubMed]
- Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat Rev Immunol 2011;11:750-61. [Crossref] [PubMed]
- Kim E, Um H, Park J, et al. TM4SF5-dependent crosstalk between hepatocytes and macrophages to reprogram the inflammatory environment. Cell Rep 2021;37:110018. [Crossref] [PubMed]
- Tsai CF, Chen JH, Yeh WL. Pulmonary fibroblasts-secreted CXCL10 polarizes alveolar macrophages under pro-inflammatory stimuli. Toxicol Appl Pharmacol 2019;380:114698. [Crossref] [PubMed]


