
Identifying and validating key genes mediating intracranial aneurysm rupture using weighted correlation network analysis and exploration of personalized treatment
Introduction
Intracranial aneurysms (IA) are a group of vasculogenic diseases caused by abnormal dilatation of the arterial wall due to damage of the elastic membrane and disruption of the media within the arterial wall resulting in a reduction of local structural integrity. The pathological process of IA formation, progression, and rupture is still unknown. According to current studies, immune inflammation plays an important role in the molecular pathology of IA (1,2), where various risk factors cause accumulation of immune cells such as macrophages and immune-related molecules leading to intracranial vascular endothelial damage and extracellular matrix (ECM) disruption and remodeling (3). This leads to dysregulation of the local vascular microenvironment and triggers activation and cascade amplification of inflammation in the vessel wall. To date, the role of immune cells and immune-related pathways and molecules including on IA has only been partially discovered. The current novel therapeutic modalities of molecular targeted therapy combined with drug scaffold technology offer new hope for the advancement of IA treatment. In animal experiments, local immunomodulatory therapies based on various cellular or molecular targets have been shown to effectively promote neurovascular lesion repair (4-6). With these explorations and developments regarding the physiopathological mechanisms of IA, there is a deeper need for potential molecular targets for IA as well as new modalities of treatment.
Aneurysmal subarachnoid hemorrhage (aSAH) caused by IA rupture is now classified as a subtype of stroke, although SAH accounts for only 3% of all stroke subtypes. It is also more rapidly progressive and intracranially destructive. However, the morbidity and mortality of aSAH are considerable (7), with an average age of onset of approximately 50 years, the incidence of IA rupture occurring approximately 9.1 times per 100,000 per year, and 40% of patients dying within the first few days to weeks after aSAH. Even after treatment, more than 46% of survivors experience disability or long-term cognitive impairment due to multiple complications (8), and only 5% of patients will resume a life not significantly different to that before the onset (9). Since cerebral hemorrhage after IA rupture is a major cause of poor patient prognosis (10), yet few clinical symptoms precede IA rupture, early and accurate screening for pre-rupture IA can provide patients with more time for anticipatory treatment.
The main clinical screening tests for IA are computed tomography angiography (CTA) and magnetic resonance angiography (MRA), which are non-invasive tests that require three-dimensional (3D) modeling to accurately depict the morphology of the cerebral blood vessels, yet CTA and MRA are more time-consuming. The gold standard for the diagnosis of IA is cerebral angiography, which requires an experienced clinician to perform, despite some discoveries of diagnostic markers in IA, their exploration has been limited. Therefore, there is a need to further search for valuable biomarkers that can be used for IA screening in a convenient and inexpensive manner.
To address these considerations, we used the GSE13353, GSE15629, and GSE54083 datasets from the Gene Expression Omnibus (GEO) database, which include 19 unruptured intracranial aneurysms (UIAs) and 27 tissue samples of ruptured intracranial aneurysms (RIAs), to construct and validate potential diagnostic marker genes for RIAs. We further explored the relationship between these diagnostic markers and immune cells, finally we also used the Connectivity Map (CMap) database to explore possible small molecule drugs for the treatment of RIAs and used molecular docking to validate the reliability of these drugs in relation to the diagnostic genes. Overall, this study characterizes the reliability of the diagnosis of IA and the development of related drugs, providing attractive genetic signatures for ancillary diagnosis and rupture prediction, contributing to mechanistic understanding and accurate diagnosis and treatment of IA. We present the following article in accordance with the STREGA reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4068/rc).
Methods
Data download
We performed a search for IAs through the GEO database and selected data on RIAs and UIAs by filtering and downloading GSE13353 (containing a total of 11 RIA and 8 UIA specimens), GSE15629 (which contained 8 ruptured and 6 human UIAs and 5 RIAs), and GSE54083 (gene expression levels were determined in 8 RIAs, 5 UIAs, and 10 superficial temporal arteries), and downloaded the relevant platform data separately for subsequent analysis. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Data processing and DEG screening
We first annotated the above downloaded data using perl, then normalized and merged the annotated GSE13353 and GSE15629 data using the limma and sva packages of R software (The R Foundation For Statistical Computing, Vienna, Austria), and finally used the limma package for screening of differentially expressed genes (DEGs) in ruptured aneurysms with the following screening criteria: log fold change (FC) ≥1, adjusted P value <0.05.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of DEGs
We took the obtained differentially expressed ruptured aneurysm genes and visualized the results using the R software packages clusterProfiler and org.Hs.eg.db for GO and KEGG enrichment analysis of clusters (11) and the ggplot2 package for visualization of the results. All DEGs were classified into three categories: biological process (BP), cellular component (CC), and molecular function (MF). The KEGG (https://www.kegg.jp/kegg/pathway.html) was used to predict the metabolic pathway classification and the results were presented using the ggplot2 package.
Protein-protein interaction (PPI) network construction
The Search Tool for the Retrieval of Interacting Genes/protein database (STRING; https://string-db.org/) was used to obtain interaction information for proteins with a combination score >0.4, which is 0.4. The CytoNCA plugin in Cytoscape software (version 3.8.2; https://cytoscape.org/) was used to visualize the network.
Weighted co-expression network construction
We used the weighted gene co-expression network analysis (WGCNA) R package (12) to analyze normalized data for GSE13353, GSE15629 combined to construct co-expression networks of RIA samples and UIA samples. We selected the 25% of genes with the highest variance, and the 2 WGCNA analyses involved 19 RIA samples and 19 UIA samples, respectively. Neighbor-joining matrices storing the information of the whole co-expression network were built based on Pearson correlation matrix. A topological overlap measure matrix was obtained from the adjacency matrix to detect the gene modules of the co-expression network. The WGCNA converted the adjacency matrix into a topological overlap matrix (TOM) based on a soft threshold power. Subsequently, the genes were divided into different modules using the TOM-based phase dissimilarity measure. We set the minModuleSize and mergeCutHeight cutoff thresholds to 30 and 0.25, respectively.
Screening of diagnostic genes
We took the intersection of the most relevant modular genes and DEGs with clinical features, combined them with GSE13353, GSE15629 for normalized data, constructed least absolute shrinkage and selection operator (LASSO) models using the glmnetR package, and validated the diagnostic models using GSE54083 annotated post data. The accuracy of the models was validated separately using the pROC package.
Single-sample gene set enrichment analysis (ssGSEA) single gene and immune infiltration analysis
To understand the immune microenvironment of RIAs, we quantified immune infiltration and associated immune functions by single-sample gene set enrichment analysis (ssGSEA), which calculates an enrichment score representing the level of immune cell infiltration and immune-related pathway activity (13). The ggplot2 package was also used to draw a small tilde map to depict the distribution of immune cells and their differences.
Gene set enrichment analysis (GSEA)
In order to understand the impact of RIA diagnostic genes on disease development mechanisms, we performed a ssGSEA, using the limma package, org.Hs.eg.db, clusterProfiler package (14), and GSEA of diagnostic genes, which were then visualized.
Screening for potential small molecule drugs and molecular docking
The CMap database (https://portals.broadinstitute.org/cmap/) can discover functional associations between relevant drugs, genes, and diseases through common gene expression changes (15). First, we divided DEGs and into up-regulated and down-regulated groups. They were uploaded to CMap for GSEA: those with enrichment values close to −1 were considered more likely to reverse the biological state of RIAs, and results with a P value <0.05 were considered statistically significant. The two-dimensional (2D) structures of potential active ingredients were downloaded from the PubChem (https://pubchem.ncbi.nlm.nih.gov/) and the Protein Data Bank (PDB; https://www.rcsb.org/) databases, respectively. Then, molecular docking of core targets to small molecule drugs was completed using autoDOCK software analysis (16). Their binding energies were visualized with a heat map, the lower the binding energy, the better the docking effect. Finally, as in our previous work, the results were visualized for the molecular pair with the lowest binding energy of the drug small molecule to the protein macromolecule to demonstrate their optimal interaction sites (17).
Statistical analysis
All statistical analyses were performed using R version 4.1.0. Statistical significance was set at P<0.05.
Results
Study process
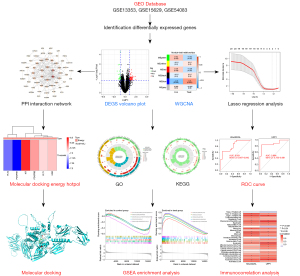
This study was sequentially divided into three phases. Figure 1 illustrates all the processes analyzed. Firstly, the IA sequencing data from the GEO database was searched and DEGs were screened, and GO and KEGG analyses were performed on these targets. Next, the WGCN was constructed using the IA GSE13353 and GSE15629 datasets, and the DEGs were intersected with the clinical module genes. Then, the intersected genes were used for the construction of the diagnostic model, and the diagnostic model was validated using GSE54083. Finally, the diagnostic genes were analyzed for immune infiltration, and the relationship between the diagnostic genes and immune infiltration and immune cells was investigated.
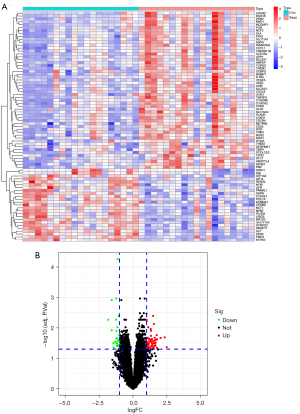
Identification of DEGs in RIAs
Using the limma package of R software to normalize and merge the downloaded data for GSE13353, GSE15629 data and to screen for DEG analysis, a total of 79 genes were determined to be significantly differentially expressed in RIAs compared with normal UIA samples, including 54 upregulated and 25 downregulated genes. Based on the ploidy changes, PPBP and CXCL8 were identified as the top 2 most up-regulated genes, and ATP1A2 and FMO2 were the top 2 most down-regulated genes. According to the screening criteria: fdrFilter <0.05, logFCfilter >1, a heat map (Figure 2A) and a volcano map (Figure 2B) were plotted using the limma software package to visualize the results of differential gene expression.
GO and KEGG enrichment analysis of RIA DEGs
We further analyzed 79 DEGs of RIAs for GO and KEGG enrichment using R software (Figure 3). The specific results of GO functional enrichment included analysis of BP, CC, and MF. The GO analysis showed that the 79 targets were widely distributed, with BP mainly focused on neutrophil activation, neutrophil degranulation, neutrophil activation involved in immune response, neutrophil-mediated immunity, and positive regulation of angiogenesis, among others. The CC were mainly distributed in platelet alpha granule lumen and specific granule, for which CC is mainly involved in virus receptor activity, exogenous protein binding, cytokine binding, and immune receptor activity, among others.
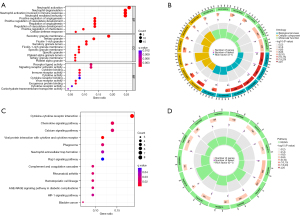
In addition, these 79 targets are also involved in multiple signaling pathways, such as cytokine-cytokine receptor interaction, chemokine signaling pathway, complement and coagulation cascades, calcium signaling pathway, HIF-1 signaling pathway, neutrophil extracellular trap formation, and Rap1 signaling pathway, among others.
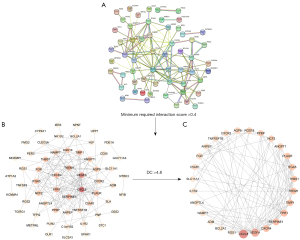
Potential core proteins of RIAs
A PPI network was performed on 79 DEGs of RIAs (Figure 4A), including 58 nodes and 140 edges (Figure 4B). Then, a subnet consisting of 27 nodes and 103 edges was obtained using the CytoNCA plugin for modular analysis (Figure 4C). We considered these 27 final genes the core genes for RIAs.
Results of WGCNA of RIA samples
We first merged and performed sample clustering analysis of the GSE13353 and GSE15629 datasets, then set the correlation soft threshold, and obtained 9 clustering_modules by calculation, and finally obtained 6 module_traits, in which the expression correlation of blue module with ruptured aneurysm region reached 0.5, (P=0.001). This module had 36 co-expressed genes, and after finally analyzing the co-expressed genes with DEGs, we identified 24 common genes (Figure 5).
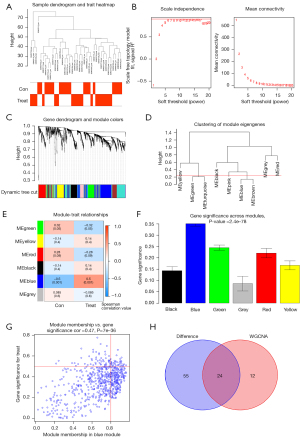
Two-gene based logistic regression model
We used LASSO regression for the 24 DEGs included in the above analysis (Figure 6A,6B), using their expression values and sample type (RIA vs. normal) as continuous predictor variables and categorical response values, respectively. We obtained the genes with the lowest crossover error by crossover analysis, which finally yielded 2 genes, UPP1 and KIAA0226L, with P values <0.05 in the logistic regression model. In addition to the healthy control samples, the expression levels of UPP1 and KIAA0226L in RIA samples were high. Therefore, we finally constructed a logistic regression model based on UPP1 and KIAA0226L. The performance of the model to differentiate RIA from normal samples was evaluated by 5-fold cross-validation. The accuracy of the model in the 5-fold cross-validation was assessed using the area under the curve (AUC) values. As a result, the AUC ranged from 0.78 to 1, with a mean AUC of 0.92, indicating the reliability of the model in distinguishing RIA from normal samples (Figure 6C,6D).
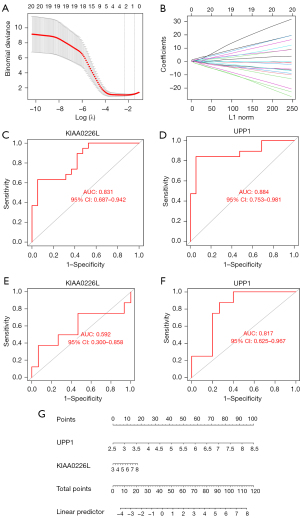
In addition, the performance of the logistic regression model based on the UPP1 and KIAA0226L genes was evaluated in another dataset, GSE54083, which included 8 RIA samples and 15 UIA samples. The results showed that the AUC value of KIAA0226L in dataset GSE5408 was 0.592 and the AUC value of UPP1 was 0.817, indicating the reliability of our model (Figure 6E,6F), and the nomogram we used to provide a score for the diagnosis of RIA (Figure 6G).
Immunocorrelation analysis of RIA tissue and diagnostic genes
We performed GSEA for UIAs and RIAs using the immunesigdb.gmt gene set file in order to explore the specific mechanism of action of our diagnostic genes in RIAs, after merging and homogenizing the GSE13353 and GSE15629 datasets, as shown in (Figure 7A,7B). Enriched in control group active gene sets were GSE21774 CD62L POS CD56 BRIGHT VS CD62L NEG CD56 DIM NK CELL DN, GSE22935 WT VS MYD88 KO MACROPHAGE DN,GSE2405 HEAT KILLED VS LIVE A PHAGOCYTOPHILUM STIM NEUTROPHIL 9H DN, GSE31622 WT VS KLF3 KO BCELL UP, GSE32533 WT VS MIR17 OVEREXPRESS ACT CD4 TCELL DN, and the gene sets that are active in the treatment group were GOLDRATH NAIVE VS MEMORY CD8 TCELL DN, GSE10325 BCELL VS MYELOID DN, GSE10325 CD4 TCELL VS MYELOID DN, GSE10325 LUPUS BCELL VS LUPUS MYELOID DN, GSE10325 LUPUS CD4 TCELL VS LUPUS MYELOID DN. These results suggest that RIAs may be associated with NAIVE, MEMORY CD8 TCELL, B CELL, CD4 T CELL, and other immune gene sets were associated.
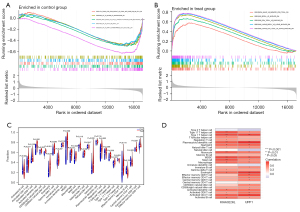
We then performed GSEA of the UPP1 and KIAA0226L genes, respectively, to explore the correlation between individual genes and immune cells, and the results are shown in (Figure 7C) indicating a correlation with activated CD4 T cell, activated dendritic cell, CD56 dim natural killer cell, eosinophil, gamma delta T cell, myeloid-derived suppressor cell (MDSC), macrophage, mast cell, natural killer T cell, neutrophil, regulatory T cell, type 1 T helper cell, type 17 T helper cell, and so on. In addition, we performed immune cell correlation analysis for diagnostic genes separately (Figure 7D). Our results showed that these 2 genes are associated with activated B cell, activated CD4 T cell, activated CD8 T cell, activated dendritic cell, eosinophil, macrophage, mast cell, MDSC, monocyte, natural killer T cell, neutrophil, plasmacytoid dendritic cell, type 1 T helper cell, type 17 T helper cell, and other immune cells are associated, and these immune cells may be involved in the pathophysiological alterations of IA rupture.
GSEA of RIA tissue and diagnostic genes
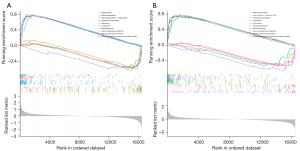
We next performed single gene GSEA enrichment analysis for UPP1 and KIAA0226L as shown in Figure 8. Interestingly, in these enrichment analyses, both were associated with vascular smooth muscle contraction and interaction of viral proteins with cytokines and cytokine receptors, and GSEA for KIAA0226L was also associated with taurine and hypotaurine metabolism. These results suggest that UPP1 and KIAA0226L may be involved in vascular smooth muscle contraction and activation of cytokines and cytokine receptors, which in turn affect the vasculature of IAs, leading to their rupture.
Exploration of potential small molecule drugs for the treatment of RIAs
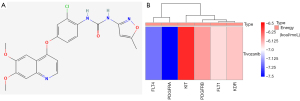
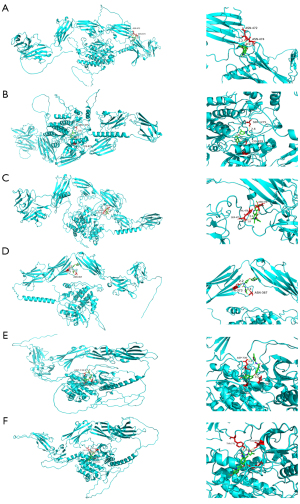
In order to explore potential small molecule drugs for the treatment of RIAs, we downloaded the results of the differentially expressed up- and down-regulated genes through the CMap database and selected a score <−85%, for which there are 5 MEK inhibitors and perturbagens, and we obtained a total of 23 inhibitors (Table 1). Tivozanib is a vascular endothelial growth factor receptor (VEGFR) inhibitor, and among the differential proteins identified, FLT1, FLT4, KDR, KIT, PDGFRA, PDGFRB, and others were enriched on VEGFR inhibitor. Therefore, we performed molecular docking of the 6 abovementioned proteins with tivozanib respectively (see Figure 9A for the 2D structure of tivozanib small molecule, and Figure 9B for the docking energy heat map). We found that the energy required for docking between PDGFRA and FLT4 was the lowest. We observed the docking results separately (Figure 10A-10F), and the FLT1 and tivozanib AutoDock data showed that the FLT1 protein formed 2 ionic bonds at positions 472 and 474 of asparagine, respectively. The FLT4 protein formed 2 ionic bonds with tivozanib small molecule at positions 879 and 1,275 for lysine and serine, respectively. The KDR protein formed 2 ionic bonds with tivozanib small molecule at positions 879 and 1,275 for lysine and serine, respectively. The FLT4 protein formed 2 ionic bonds with tivozanib small molecule at positions 475, 476, and 478 for serine, valine, and asparagine and formed 2 covalent bonds and 1 van der Waals force with tivozanib small molecule. The tivozanib small molecule formed a van der Waals force with the asparagine at position 367 and a hydrogen bond with the tyrosine at position 373 on the KIT protein. The tivozanib small molecule formed hydrogen bonds, ionic bonds, and van der Waals forces with asparagine at positions 730, 842, and 846 on the PDGFRA protein. Tivozanib small molecule formed hydrogen bonds with leucine at position 606, a hydrogen bond and an ionic bond with tyrosine at position 771, and an ionic bond with aspartic acid at position 849 on the PDGFRB protein.
Table 1
| MOA | Perturbagen count | Perturbagen ID | Name |
|---|---|---|---|
| MEK inhibitor | 5 | BRD-K89014967, BRD-K49865102, BRD-K05104363, BRD-K57080016, BRD-K62810658 | AS-703026, PD-0325901, PD-184352, selumetinib, PD-98059 |
| p38 MAPK inhibitor | 3 | BRD-K77133231, BRD-K54330070, BRD-K52751261 | PD-169316, SB-202190, TAK-715 |
| HSP inhibitor | 2 | BRD-K51967704, BRD-K41859756 | BIIB021, NVP-AUY922 |
| FLT3 inhibitor | 2 | BRD-K12502280, BRD-K23192422 | TG-101348, lestaurtinib |
| SRC inhibitor | 2 | BRD-K47598052, BRD-K19540840 | PP-1, saracatinib |
| ABL inhibitor | 2 | BRD-K41337261, BRD-K99964838 | ZM-306416, bosutinib |
| RAF inhibitor | 2 | BRD-K01253243, BRD-K30677119 | SB-590885, PP-30 |
| IKK inhibitor | 1 | BRD-K51575138 | TPCA-1 |
| Phenylalanyl tRNA synthetase inhibitor | 1 | BRD-K39944607 | Ochratoxin-a |
| JNK inhibitor | 1 | BRD-K84085265 | CG-930 |
| VEGFR inhibitor | 1 | BRD-K53414658 | tivozanib |
| MTOR inhibitor | 1 | BRD-K67566344 | KU-0063794 |
| Glycogen synthase kinase inhibitor | 1 | BRD-K94176593 | TWS-119 |
| HMGCR inhibitor | 1 | BRD-U88459701 | Atorvastatin |
| Gonadotropin releasing factor hormone receptor agonist | 1 | BRD-A62434282 | Goserelin |
| Histone lysine methyltransferase inhibitor | 1 | BRD-K74236984 | UNC-0321 |
| Calcium channel blocker | 1 | BRD-K49404994 | Levetiracetam |
| Chelating agent | 1 | BRD-K07265709 | Razoxane |
| TGF beta receptor inhibitor | 1 | BRD-K06234293 | LY-364947 |
| Cyclooxygenase inhibitor | 1 | BRD-A22844106 | Tenoxicam |
| PI3K inhibitor | 1 | BRD-K06750613 | GSK-1059615 |
| BCR-ABL kinase inhibitor | 1 | BRD-K49328571 | Dasatinib |
| CHK inhibitor | 1 | BRD-K46056750 | AZD-7762 |
DEGs, differentially expressed genes; RIA, ruptured intracranial aneurysm; MOA, CMap mode of action.
Discussion
The prevalence of IA is estimated to be as high as 10% according to epidemiological surveys. In general, medium-sized and small IA are asymptomatic prior to rupture. Although a majority (91.5%) of asymptomatic and untreated IA prove to be stable and do not continue to grow, periodic imaging is required for those untreated aneurysms. Maximum caution and care need to be exercised in the treatment of growing but still asymptomatic aneurysms, which are found to rupture in 4% of patients; however, patients with IA experience severe SAH in the event of IA rupture (18,19). Among the subtypes of stroke, SAH is the most devastating and causes early damage to the cerebral nerves leading to severe neurological deficits and even death at early onset (20,21). The current treatment for IA consists of minimally invasive endovascular embolization and open surgical resection of cerebral aneurysms. The aim of both methods is to isolate the aneurysm from the circulation to prevent rupture. However, antiplatelet and anticoagulant agents are generally required after endovascular embolization, and the role of these in IAs has been controversial. Patients on long-term aspirin have a reduced risk of rupture, and the use of antiplatelet aggregating agents such as aspirin may be associated with SAH (22,23). The current most effective treatment for vasospasm due to SAH is the calcium channel antagonist-nimodipine, which has a single selective treatment. Therefore, identifying the potential key pathways, genes, and associated potential small molecule agents in the process of IA the role in reversing rupture is an urgent research task.
In this study, multiple datasets were used to identify deg associated with IA rupture for WGCNA. Gene modules associated with IA rupture were also identified using WGCNA. The resultant module gene was selected as the blue module most associated with UIA, and the blue module was intersected with the deg to identify a total of 24 core genes. These results suggest that these 24 genes may be the most critical genes for the formation and development of IA. These common genes were used for drug discovery through the CAMP database. The next step was to construct a model for the diagnosis, using receiver operating characteristic (ROC) curves to validate the diagnostic efficacy, and on the other hand the performance of a logistic regression model based on the UPP1 and KIAA0226L genes was assessed using the GSE54083, ROC curves to validate the reliability of the UPP1 and KIAA0226L diagnosis, and a nomogram to calculate the probability of RIA occurrence. To investigate the mechanism of IA rupture, we performed immune-related GSEA on our samples, and the results suggested that RIAs might be associated with immune cells such as naive, memory CD8 T cell, B cell, and CD4 T cell. A single-cell sequencing analysis of aneurysm wall-derived T cells in a pediatric subarachnoid hemorrhage case revealed that naïve T cells and typical memory T cells were mainly confined to peripheral blood, whereas CD4-CD8 T cells were strongly enriched in the IA wall. In contrast, immune inflammation is now considered an important pathogenic factor in the formation and rupture of IA and in the development of complications after endovascular treatment (24-26). This also provides a new direction to further investigate the pathological development of immune cells in the walls of ruptured IA vessels as well as new therapeutic targets, which is a new promising research direction for the treatment of our IAs.
After merging and sample clustering analysis of the GSE13353 and GSE15629 datasets with 79 DEGs of RIAs to take intersections, we obtained 24 intersecting genes, and we finally searched for 2 diagnostic signature genes by LASSO regression, 1 of which is KIAA0226L, which is a regulator of autophagy that promotes autophagosome maturation by facilitating the biogenesis of phosphatidylinositol 3-phosphate (PtdIns3P) in late steps of autophagy (27). It has been shown that cellular autophagy plays an important role in many neurological disorders, including brain injury and secondary brain edema due to brain hemorrhage (28,29). On the one hand, resveratrol protects early brain injury after subarachnoid hemorrhage by activating autophagy and inhibiting apoptosis mediated by the Akt/mTOR pathway (30). Then IA formation and its rupture may be associated with the expression of certain autophagic proteins, and activation of phosphatidylinositol 3-kinase/protein kinase B pathway by a vanadyl compound mediates its neuroprotective effect in mouse brain ischemia (31). On the other hand: activation of the phosphatidylinositol 3-kinase-Akt pathway, combined with translocation of p47phox, p22phox and rac1, contributes to high glucose-induced superoxide production, leading to impairment of adenosine triphosphate (ATP)-sensitive K-channel function in human visceral arterial smooth muscle cells (32). As for the blood-brain barrier (BBB), loss of tight junction (TJ) proteins in brain endothelial cells (ECs) leads to BBB disruption, and intracerebroventricular injection of autophagy inhibitors rescues BBB leakage induced by GM130 silencing, which protects TJ from damage by maintaining proper autophagy. We suggest that GM130-regulated selective autophagy regulates BBB integrity and GM130 upregulation inhibits the autophagy-lysosome pathway, which may maintain BBB function (33). Our GSEA analysis of KIAA0226L showed that its down-regulated expression was associated with vascular smooth muscle contraction. In contrast, a study showed that miR-331-3p is a direct negative regulator of TNF-α and CD14, and its elevation also reduced apoptosis and prevented the synthetic phenotype of vascular smooth muscle cells (VSMC) and increased their proliferation (34). Therefore, we may in the future exert a protective effect on intracranial aneurysmal vessels by inhibiting the protein expression of KIAA0226L, thereby reducing the occurrence of endothelial autophagy as well as reversing VSMC dysfunction, ultimately.
The other diagnostic gene was UPP1, which catalyzes the reversible phosphorylytic cleavage of uridine and deoxyuridine to uracil and ribose- or deoxyribose-1-phosphate (dRP) (35). The produced molecules are then utilized as carbon and energy sources or in the rescue of pyrimidine bases for nucleotide synthesis, and dRP has an important role in angiogenesis, dRP is a proangiogenic paracrine stimulus released by cancer cells, platelets, and macrophages and acting on ECs, thereby directly activating NADPH oxidase 2 to trigger nuclear factor kappa B-dependent angiogenesis, and platelet-derived dRP stimulates EC migration by upregulating integrin β (3) in a reactive oxygen species-dependent manner (36,37). Interestingly GSEA of UPP1 showed that its downregulated expression was also associated with vascular smooth muscle contraction. These studies show that IA can stimulate the expression of UPP1 and thus accelerate the production of dRP, which in turn promotes the growth of ECs and exerts a certain effect on the healing of blood vessels after IA rupture, thus providing some ideas for the development of target-specific drugs for IA patients.
Important clinical risk factors for IA include being female, hypertension and smoking. Familial intracranial aneurysms (FIAs), on the other hand, are found in approximately 6-20% of patients with IAs, suggesting that genetic susceptibility may play a role in their pathogenesis. One study used whole exome sequencing (WES) to identify possible IA-associated variants, sequencing 13 individuals from three families, seven of whom had IA. They identified (PLOD3 c.1315G>A, NTM c.968C>T and CHST14 c.58C>T) as any significant copy number variants in the family (38). In one other study, based on chromosomal data from genome-wide association studies (GWAS) of 250 patients diagnosed with IA and 296 controls, our fine mapping analysis showed that four functional candidate variants, GBA, TCF24, OLFML2A and ARHGAP32, were associated with susceptibility and pathogenesis of IA (39). and the presence of the T allele in the rs12976445 polymorphism was associated with a lower risk of IA rebleeding (40). However, the results obtained in these studies are limited by the small sample sizes used and further research is needed.
Digital subtraction angiography (DSA) is currently one of the best methods for diagnosing IAs because it is invasive and does not show occluded distal vessels, in addition to its high cost and potential risks. DSA is therefore not suitable for early screening and treatment of IAs. Recently, detection of microRNA (miRNA) in circulating immune cells has been used as a means of screening for diagnostic markers of vascular diseases such as IAs. One study used serum from patients with IA to examine cytokines to analyse serum cytokine concentrations. Clinical data and cytokines were compared between groups. The study has found elevated levels of IL-15, MCP-1 and TNF-β in UIAs (41). On the other hand, one study used 66 patients with UIAs, 69 patients with RA, and 68 patients with hydrocephalus to perform real-time polymerase reaction to detect cerebrospinal fluid exosome miR-152-3p. exosome miR-152-3p was lower in patients with IAs, with RA being lower than in patients with UA (<0.001). the ROC confirmed that exosome miR-152-3p not only differentiated between UA patients and patients with hydrocephalus, but also predicted SAH in patients with IAs (42). These studies provide us with some new ideas as well as a more diverse approach to diagnosing IA, which also needs to be studied on a larger scale.
Currently, no prophylactic treatment or imaging follow-up is the most effective strategy for patients with aneurysms 3 mm or smaller, resulting in better health outcomes. More aggressive imaging surveillance or prophylactic treatment for aneurysm growth should be reserved for patients at high risk of rupture (43). The main treatment options for large or RIAs include conservative treatment combined with imaging follow-up, surgical clamping therapy, and endovascular interventions, and the current pharmacological treatment for IAs is relatively limited in terms of its development due to the fact that, after recent experimental studies, IA is considered a chronic inflammatory disease affecting intracranial arteries. For example, neutrophils are able to promote IA rupture through extracellular traps development (44), which suggests the potential of anti-inflammatory drugs as therapeutic agents for the treatment of IA. Whereas a cross-sectional study by a research organization showed that statins, calcium channel blockers, and angiotensin II receptor blockers were inversely associated with RIA, only non-aspirin non-steroidal anti-inflammatory drugs (NSAIDs) were positively associated with RIA (45). One study showed that as inflammatory smooth muscle cells (iSMCs) in cerebral aneurysms convert to ECs, the cells have reduced contractility and secrete more matrix metalloproteinases, and iSMCs induce collagen denaturation, which induces EC escape. In addition, iSMCs induce EC dysfunction. Cellular escape and dysfunction expose the vascular mesangium to blood flow, which ultimately attenuates inflammation and slows aneurysm development (46).
A recent study of the effect of drug inhibition of epidermal growth factor receptor (EGFR) on IA occurrence showed that activation of EGFR in rat VSMCs and vascular remodeling and pro-inflammatory transformation in rat VSMCs were reduced by treatment with erlotinib. In vitro experiments also revealed that EGFR activation in human brain VSMCs can be induced by TNF-α and that the EGFR inhibitor erlotinib can attenuate the incidence of IA (47), which may also make EGFR inhibitors a new targeted agent to slow the progression of IAs. Therefore, in our search for novel targets and corresponding drugs for IAs, 79 differential genes expressing up- and down-regulation of ruptured aneurysms were imported into the CMap database, and we 23 eventually we found tivozanib to be a VEGFR inhibitor, and the enriched proteins were FLT1, FLT4, KDR, KIT, and PDGFRA. A prospective study found that serum FLT1 levels were elevated in aSAH patients at risk of severe vasospasm (48), and an experimental study found that circLIFR and KDR were downregulated and miR-1299 was upregulated in arterial wall tissue and human umbilical artery smooth muscle cells from IA patients. Forced expression of circLIFR enhanced human umbilical arterial smooth muscle cells proliferation, migration, invasion, and prevention of apoptosis (49). In contrast, platelet-derived growth factor receptor β (PDGFRB) is currently expressed and mutated in intracranial, coronary, aortic, and radial artery vascular bed aneurysms (50,51). Thus, the 6 VEGFR inhibitor-related proteins mentioned above may be able to be targets for the treatment of IAs, and our molecular docking showed that tivozanib binds to KDR and PDGFRB. Tivozanib small molecule drug was approved for use by the Food and Drug Authority (FDA) in 2021 for the treatment of relapsed or refractory renal cell carcinoma, and it has the ability to reduce chemotherapy-induced myelosuppressive effects (52). Tivozanib can reduce the accumulation of Tregs and myeloid-derived suppressor cells by inhibiting the c-Kit/SCF axis, which is currently found in the peripheral blood of IA patients with Th17/Treg imbalance, and some investigators have inhibited the PI3k/Akt/NF-κB through the bone marrow mesenchymal stem cell-derived exosome has-miR-23b-3p signaling pathway to target KLF5 to maintain Th17/Treg homeostasis, thereby inhibiting IA formation (53-55). Tivozanib may become a more promising VEGFR inhibitor to delay the progression of IA, which needs further investigation.
In this study, we first obtained more reliable diagnostic genes KIAA0226L and UPP1 by LASSO regression model, and the AUC values of ROC curves reached 0.833 and 0.844, respectively. We finally visualised the nomogram for the UPP1 and KIAA0226L genes and we were able to calculate our scores from the expression of the UPP1 and KIAA0226L genes to obtain the final score for the diagnosis. This is used to calculate the probability of RIA. Their GSEA analysis showed that the down-regulated expression of KIAA0226L and UPP1 was associated with vascular smooth muscle contraction. Moreover, the immune cell contents of activated B cell, activated CD4 T cell, and activated CD8 T cell were positively correlated with KIAA0226L and UPP1. We also found 23 inhibitors for the treatment of ruptured aneurysms through the CMap database. Among them, VEGFR inhibitors may slow down the progression of a ruptured aneurysm, and the enriched proteins are FLT1, FLT4, KDR, KIT, PDGFRA, and PDGFRB. Tivozanib is a VEGFR inhibitor, and we also further explored the targets of their interaction by molecular docking.
his study had some limitations and further in-depth study is required. First, only the GSE13353, GSE15629, and GSE54083 datasets were computerized to explore diagnostic genes as well as possible markers for treatment and drugs. Next, we need to further confirm the therapeutic effect of tivozanib on IA through basic experiments using the core small molecule drugs in our conclusion. In addition, clinical trials of tivozanib for the treatment of patients with IA were not performed. In summary, through a comprehensive systematic analysis, we obtained KIAA0226L and UPP1 as genes for the diagnosis of IA rupture and are hopeful that tivozanib will be a novel and promising agent for targeting and mitigating the progression of IA.
Conclusions
Through comprehensive and systematic analysis of the IA dataset, we finally ascertained that KIAA0226L and UPP1 are a novel gene for the diagnosis of IA rupture, and our immune correlation analysis suggested that activated B cells, activated CD4 T cells, activated CD8 T cell, and other immune cell contents were positively correlated with KIAA0226L and UPP1. We also explored tivozanib as a VEGFR inhibitor, which is anticipated to become a novel drug to delay the progression of IA.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-4068/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-4068/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Etminan N, Rinkel GJ. Unruptured intracranial aneurysms: development, rupture and preventive management. Nat Rev Neurol 2016;12:699-713. [Crossref] [PubMed]
- Zhang Zeyu, Fang Yuanjian, Lenahan Cameron, et al. The role of immune inflammation in aneurysmal subarachnoid hemorrhage. Exp Neurol 2021;336:113535. [Crossref] [PubMed]
- Kanematsu Y, Kanematsu M, Kurihara C, et al. Critical roles of macrophages in the formation of intracranial aneurysm. Stroke 2011;42:173-8. [Crossref] [PubMed]
- Zhang H, Watanabe R, Berry GJ, et al. Inhibition of JAK-STAT Signaling Suppresses Pathogenic Immune Responses in Medium and Large Vessel Vasculitis. Circulation 2018;137:1934-48. [Crossref] [PubMed]
- Cortese J, Rasser C, Even G, et al. CD31 Mimetic Coating Enhances Flow Diverting Stent Integration into the Arterial Wall Promoting Aneurysm Healing. Stroke 2021;52:677-86. [Crossref] [PubMed]
- Liu Y, Liu P, Song Y, et al. A heparin-rosuvastatin-loaded P(LLA-CL) nanofiber-covered stent inhibits inflammatory smooth-muscle cell viability to reduce in-stent stenosis and thrombosis. J Nanobiotechnology 2021;19:123. [Crossref] [PubMed]
- Tawk RG, Hasan TF, D'Souza CE, et al. Diagnosis and Treatment of Unruptured Intracranial Aneurysms and Aneurysmal Subarachnoid Hemorrhage. Mayo Clin Proc 2021;96:1970-2000. [Crossref] [PubMed]
- Lublinsky S, Major S, Kola V, et al. Early blood-brain barrier dysfunction predicts neurological outcome following aneurysmal subarachnoid hemorrhage. EBioMedicine 2019;43:460-72. [Crossref] [PubMed]
- Passier PE, Visser-Meily JM, van Zandvoort MJ, et al. Prevalence and determinants of cognitive complaints after aneurysmal subarachnoid hemorrhage. Cerebrovasc Dis 2010;29:557-63. [Crossref] [PubMed]
- Vergouwen MD, Jong-Tjien-Fa AV, Algra A, et al. Time trends in causes of death after aneurysmal subarachnoid hemorrhage: A hospital-based study. Neurology 2016;86:59-63. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. [Crossref] [PubMed]
- Lamb J, Crawford ED, Peck D, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 2006;313:1929-35. [Crossref] [PubMed]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31:455-61. [PubMed]
- Wu J, Li XY, Liang J, et al. Network pharmacological analysis of active components of Tongqiao Huoxue Decoction in the treatment of intracerebral hemorrhage. Ann Transl Med 2022;10:567. [Crossref] [PubMed]
- Moon J, Cho YD, Yoo DH, et al. Growth of Asymptomatic Intracranial Fusiform Aneurysms: Incidence and Risk Factors. Clin Neuroradiol 2019;29:717-23. [Crossref] [PubMed]
- van der Kamp LT, Rinkel GJE, Verbaan D, et al. Risk of Rupture After Intracranial Aneurysm Growth. JAMA Neurol 2021;78:1228-35. [Crossref] [PubMed]
- Cheng C, Jiang L, Yang Y, et al. Effect of APOE Gene Polymorphism on Early Cerebral Perfusion After Aneurysmal Subarachnoid Hemorrhage. Transl Stroke Res 2015;6:446-50. [Crossref] [PubMed]
- Etminan N. Aneurysmal subarachnoid hemorrhage--status quo and perspective. Transl Stroke Res 2015;6:167-70. [Crossref] [PubMed]
- Toussaint LG 3rd, Friedman JA, Wijdicks EF, et al. Influence of aspirin on outcome following aneurysmal subarachnoid hemorrhage. J Neurosurg 2004;101:921-5. [Crossref] [PubMed]
- Schmidt M, Johansen MB, Lash TL, et al. Antiplatelet drugs and risk of subarachnoid hemorrhage: a population-based case-control study. J Thromb Haemost 2010;8:1468-74. [Crossref] [PubMed]
- Berge J, Tourdias T, Moreau JF, et al. Perianeurysmal brain inflammation after flow-diversion treatment. AJNR Am J Neuroradiol 2011;32:1930-4. [Crossref] [PubMed]
- van der Poll T, Shankar-Hari M, Wiersinga WJ. The immunology of sepsis. Immunity 2021;54:2450-64. [Crossref] [PubMed]
- Moschetti G, Vasco C, Clemente F, et al. Deep Phenotyping of T-Cells Derived From the Aneurysm Wall in a Pediatric Case of Subarachnoid Hemorrhage. Front Immunol 2022;13:866558. [Crossref] [PubMed]
- Cheng X, Ma X, Ding X, et al. Pacer Mediates the Function of Class III PI3K and HOPS Complexes in Autophagosome Maturation by Engaging Stx17. Mol Cell 2017;65:1029-1043.e5. [Crossref] [PubMed]
- He Y, Wan S, Hua Y, et al. Autophagy after experimental intracerebral hemorrhage. J Cereb Blood Flow Metab 2008;28:897-905. [Crossref] [PubMed]
- Durocher M, Knepp B, Yee A, et al. Molecular Correlates of Hemorrhage and Edema Volumes Following Human Intracerebral Hemorrhage Implicate Inflammation, Autophagy, mRNA Splicing, and T Cell Receptor Signaling. Transl Stroke Res 2021;12:754-77. [Crossref] [PubMed]
- Guo D, Xie J, Zhao J, et al. Resveratrol protects early brain injury after subarachnoid hemorrhage by activating autophagy and inhibiting apoptosis mediated by the Akt/mTOR pathway. Neuroreport 2018;29:368-79. [Crossref] [PubMed]
- Shioda N, Ishigami T, Han F, et al. Activation of phosphatidylinositol 3-kinase/protein kinase B pathway by a vanadyl compound mediates its neuroprotective effect in mouse brain ischemia. Neuroscience 2007;148:221-9. [Crossref] [PubMed]
- Kinoshita H, Matsuda N, Kaba H, et al. Roles of phosphatidylinositol 3-kinase-Akt and NADPH oxidase in adenosine 5'-triphosphate-sensitive K+ channel function impaired by high glucose in the human artery. Hypertension 2008;52:507-13. [Crossref] [PubMed]
- Deng S, Hu Q, Chen X, et al. GM130 protects against blood-brain barrier disruption and brain injury after intracerebral hemorrhage by regulating autophagy formation. Exp Gerontol 2022;163:111772. [Crossref] [PubMed]
- Fan W, Liu Y, Li C, et al. microRNA-331-3p maintains the contractile type of vascular smooth muscle cells by regulating TNF-α and CD14 in intracranial aneurysm. Neuropharmacology 2020;164:107858. [Crossref] [PubMed]
- Watanabe S, Uchida T. Cloning and expression of human uridine phosphorylase. Biochem Biophys Res Commun 1995;216:265-72. [Crossref] [PubMed]
- Pula G, Garonna E, Dunn WB, et al. Paracrine stimulation of endothelial cell motility and angiogenesis by platelet-derived deoxyribose-1-phosphate. Arterioscler Thromb Vasc Biol 2010;30:2631-8. [Crossref] [PubMed]
- Vara D, Watt JM, Fortunato TM, et al. Direct Activation of NADPH Oxidase 2 by 2-Deoxyribose-1-Phosphate Triggers Nuclear Factor Kappa B-Dependent Angiogenesis. Antioxid Redox Signal 2018;28:110-30. [Crossref] [PubMed]
- Song Y, Lee JK, Lee JO, et al. Whole Exome Sequencing in Patients with Phenotypically Associated Familial Intracranial Aneurysm. Korean J Radiol 2022;23:101-11. [Crossref] [PubMed]
- Hong EP, Youn DH, Kim BJ, et al. Fine-mapping of intracranial aneurysm susceptibility based on a genome-wide association study. Sci Rep 2022;12:2717. [Crossref] [PubMed]
- Xiong W, Yao W, Gao Z, et al. Rs12976445 polymorphism is associated with the risk of post-SAH re-bleeding by modulating the expression of microRNA-125 and ET-1. Sci Rep 2022;12:2062. [Crossref] [PubMed]
- Yang S, Liu Q, Yang J, et al. Increased Levels of Serum IL-15 and TNF-β Indicate the Progression of Human Intracranial Aneurysm. Front Aging Neurosci 2022;14:903619. [Crossref] [PubMed]
- Li Y, Wu A, Dai W, et al. Cerebrospinal fluid exosomal miR-152-3p predicts the occurrence of subarachnoid haemorrhage and regulates vascular smooth muscle cell dysfunction. Folia Neuropathol 2022;60:185-94. [Crossref] [PubMed]
- Malhotra A, Wu X, Forman HP, et al. Management of Tiny Unruptured Intracranial Aneurysms: A Comparative Effectiveness Analysis. JAMA Neurol 2018;75:27-34. [Crossref] [PubMed]
- Korai M, Purcell J, Kamio Y, et al. Neutrophil Extracellular Traps Promote the Development of Intracranial Aneurysm Rupture. Hypertension 2021;77:2084-93. [Crossref] [PubMed]
- Shimizu K, Imamura H, Tani S, et al. Candidate drugs for preventive treatment of unruptured intracranial aneurysms: A cross-sectional study. PLoS One 2021;16:e0246865. [Crossref] [PubMed]
- Liu P, Shi Y, Fan Z, et al. Inflammatory Smooth Muscle Cells Induce Endothelial Cell Alterations to Influence Cerebral Aneurysm Progression via Regulation of Integrin and VEGF Expression. Cell Transplant 2019;28:713-22. [Crossref] [PubMed]
- Luo Y, Tang H, Zhang Z, et al. Pharmacological inhibition of epidermal growth factor receptor attenuates intracranial aneurysm formation by modulating the phenotype of vascular smooth muscle cells. CNS Neurosci Ther 2022;28:64-76. [Crossref] [PubMed]
- Griessenauer CJ, Chua MH, Hanafy KA, et al. Soluble Fms-Like Tyrosine Kinase 1 (sFlt-1) and Risk of Cerebral Vasospasm After Aneurysmal Subarachnoid Hemorrhage. World Neurosurg 2017;108:84-9. [Crossref] [PubMed]
- Zhang H, Zhang B, Chen C, et al. Circular RNA circLIFR regulates the proliferation, migration, invasion and apoptosis of human vascular smooth muscle cells via the miR-1299/KDR axis. Metab Brain Dis 2022;37:253-63. [Crossref] [PubMed]
- Karasozen Y, Osbun JW, Parada CA, et al. Somatic PDGFRB Activating Variants in Fusiform Cerebral Aneurysms. Am J Hum Genet 2019;104:968-76. [Crossref] [PubMed]
- Parada CA, El-Ghazali FM, Toglia D, et al. Somatic Mosaicism of a PDGFRB Activating Variant in Aneurysms of the Intracranial, Coronary, Aortic, and Radial Artery Vascular Beds. J Am Heart Assoc 2022;11:e024289. [Crossref] [PubMed]
- Roskoski R Jr. Properties of FDA-approved small molecule protein kinase inhibitors: A 2022 update. Pharmacol Res 2022;175:106037. [Crossref] [PubMed]
- Kalathil SG, Wang K, Hutson A, et al. Tivozanib mediated inhibition of c-Kit/SCF signaling on Tregs and MDSCs and reversal of tumor induced immune suppression correlates with survival of HCC patients. Oncoimmunology 2020;9:1824863. [Crossref] [PubMed]
- Sun X, Zheng X, Zhang X, et al. Exosomal microRNA-23b-3p from bone marrow mesenchymal stem cells maintains T helper/Treg balance by downregulating the PI3k/Akt/NF-κB signaling pathway in intracranial aneurysm. Brain Res Bull 2020;165:305-15. [Crossref] [PubMed]
- Song M, Jin Z, Wang P, et al. Th17/Treg imbalance in peripheral blood from patients with intracranial aneurysm. J Neurosurg Sci 2021; Epub ahead of print. [Crossref] [PubMed]
(English Language Editor: J. Jones)


