
Krüppel-like factor 5 promotes the progression of oral squamous cell carcinoma via the baculoviral IAP repeat containing 5 gene
Introduction
Oral squamous cell carcinoma (OSCC) occurs in the oral cavity including the tongue, mouth floor, soft palate, gums, and buccal mucosa (1). It is the most common tumor in the head and neck region, accounting for 90% of all oral malignancies and 1–2% of all human malignancies (2). Furthermore, 16–62% of OSCC cases arise from oral potentially malignant disorders (OPMDs) (3). OPMDs, such as oral leukoplakia, erythroplakia, and oral lichen planus (OLP), represent various lesions occurring in the oral mucosa that can progress to malignancies (4). Currently, the lack of accurate indicators for oral cancer screening in the general population has led to a high incidence of oral cancer (5). Therefore, identification of key factors that control the progression of OPMDs and OSCC is urgently required to facilitate early diagnosis and treatment of OSCC, which will improve the survival and prognosis of patients.
Krüppel-like factors (KLFs) are a family of transcription factors involved in cell proliferation, differentiation, growth, and development and are closely associated with various cancers (6). One of its family members, Krüppel-like factor 5 (KLF5), plays a critical role in the occurrence and development of multiple tumors. KLF5 regulates baculoviral inhibitor of apoptosis protein (IAP) repeat containing 5 (BIRC5) expression by interacting with protein 53 (P53) in acute lymphoblastic leukemia (7). Similarly, KLF5 inhibits BIRC5 expression by directly binding to the BIRC5 promoter in ovarian cancer stem-like cells (8). Previous studies have found that BIRC5, a member of the IAP family, is a central molecular and therapeutic target in cancer (9), and its high expression is associated with poor overall survival in patients with OSCC (10). It has been demonstrated that miR-375, as a tumor suppressor, can decrease KLF5 expression to inhibit OSCC development (11); however, the role of KLF5 in OSCC and OPMDs has not been further elucidated until now. Furthermore, the correlation between KLF5 and BIRC5 in OSCC and OPMDs remains unclear.
In this study, we investigated the differential expression of KLF5 in OSCC and OPMDs using bioinformatics analysis. Further experiments were performed to explore the association of KLF5 with the malignant biological behavior of OSCC and OPMD cells. We then analyzed the relationship between KLF5 and BIRC5 using bioinformatics analysis as well as the suppression of KLF5 expression in vitro and in vivo. We present the following article in accordance with the ARRIVE reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3728/rc).
Methods
Clinical sample acquisition
Paired OSCC, OLP, and adjacent normal mucosa samples were obtained from 2 patients pathologically diagnosed with OSCC and OLP at the Peking University School and Hospital of Stomatology (Beijing, China). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Peking University Institutional Review Board (approval No. IRB00001052–12037) and informed consent was taken from both patients. Next-generation sequencing (NGS) was used to select the differentially expressed genes, and the data are available with the Gene Expression Omnibus (GEO) series accession number GSE70666 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE 70666).
The Cancer Genome Atlas (TCGA) dataset
The mRNA expression data of 170 OSCC samples and 19 paracancerous tissues were obtained from TCGA dataset (https://www.cancer.gov/). The inclusion criteria for this study were patients diagnosed with OSCC by pathology and no history of other tumors. RNA-seq data were converted to transcripts per million values and then taken as log2 values, with the ‘scale’ function in the R package limma further applied for normalization.
Gene set enrichment analysis (GSEA)
The 170 OSCC samples were divided into high- and low-expression groups based on the best KLF5 cutoff value defined by X-tile software (version 3.6.1). To explore the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment of KLF5, the RNA expression data of both groups were extracted from the TCGA dataset. GSEA was performed using GSEA software (version 4.1.0). The predefined gene sets “c2.cp.kegg.v7.2.symbols.gmt” and “h.all.v7.5.1.symbols.gmt” were downloaded from the Molecular Signatures Database (MSigDB). Pathways and hallmarks with false discovery rate <25% and P<0.05 after performing 1,000 permutations were considered significantly enriched.
Human cell lines
OSCC cell lines (CAL27, SCC15) and the OPMD cell line (DOK) were cultured at 37 ℃ in a humidified incubator with 5% CO2. CAL27 and SCC15 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Grand Island, NY, USA) supplemented with fetal bovine serum (FBS, Gibco) and 1% penicillin/streptomycin (Solarbio, Beijing, China). DOK cells were cultured in DMEM supplemented with FBS, 1% penicillin/streptomycin, and 0.4 mM hydrocortisone (Solarbio).
Cell transfection
Cells (CAL27, SCC15, and DOK) were transfected for 48 h with small interfering RNAs (siRNAs) specifically targeting KLF5 (si-KLF5) and corresponding non-specific siRNA (si-NC) (JijiangBio, Nanjing, China) in a 6-well plate using an siRNA transfection kit (RiboBio, Guangzhou, China) according to the manufacturer’s instructions. The siRNA sequences are listed in Table 1.
Table 1
| Primer | Sense/antisense | Sequence |
|---|---|---|
| Homo-si-KLF5 | Sense | 5'-GCAGAGAUGCUCCAGAAUUTT-3' |
| Antisense | 5'-AAUUCUGGAGCAUCUCUGCTT-3' | |
| Homo-si-NC | Sense | 5'-UUCUCCGAACGUGUCACGUdTdT-3' |
| Antisense | 5'-ACGUGACACGUUCGGAGAAdTdT-3' | |
| Homo-KLF5 | Forward primer | 5'-CCTGGTCCAGACAAGATGTGA-3' |
| Reverse primer | 5'-GAACTGGTCTACGACTGAGGC-3' | |
| Homo-BIRC5 | Forward primer | 5'-CACTTTCTCCGCAGTTTCCTCA-3' |
| Reverse primer | 5'-CCAGACTTGGCCCAGTGTTTC-3' | |
| Homo-GAPDH | Forward primer | 5'-GGAGCGAGATCCCTCCAAAAT-3' |
| Reverse primer | 5'-GGCTGTTGTCATACTTCTCATGG-3' |
siRNA, small interfering RNA; qRT-PCR, quantitative real-time PCR; KLF5, Krüppel-like factor 5; NC, negative control; BIRC5, baculoviral inhibitor of apoptosis protein repeat containing 5; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Cell proliferation and apoptosis assays
Cell proliferation was evaluated after 24, 48, and 72 h using a CCK-8 assay (Dojindo, Japan). Absorbance was measured at 450 nm using a microplate reader (Bio-Rad, USA). Cells were collected and stained with Annexin V-FITC and propidium iodide (PI) following the instructions of the Annexin V-FITC Apoptosis Detection kit (Solarbio). Apoptosis was analyzed using flow cytometry (CytoFLEX, Beckman, USA).
Wound-healing cell migration assay
Cells (CAL27, SCC15, and DOK) were seeded in 6-well plates and cultured to 100% confluence. Then, a linear wound was introduced across the monolayer using a sterile 200-µL pipette tip. Non-adherent cells and debris were removed, and serum-free medium was added. Percentage wound closure (%) = (width at hour 0 – width at hour 24)/(width at hour 0) × 100. Wound areas were evaluated using ImageJ software (version 1.8.0).
Quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted and reverse-transcribed into cDNA. Subsequently, qRT-PCR was performed using primers for amplification of human KLF5, BIRC5, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The primer sequences used are listed in Table 1. All samples were analyzed in triplicate, and relative gene expression levels were quantified using the 2−ΔΔCt method.
Western blot analysis
Total protein was extracted from the cells using radioimmunoprecipitation assay lysis buffer (Solarbio) containing a protease inhibitor cocktail. Proteins (20 µg) were separated using sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE, Solarbio) and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, Massachusetts, USA). Then, membranes were incubated with primary antibodies [1:2,000 KLF5, 1:3,000 BIRC5, and 1:3,000 horseradish peroxidase (HRP)-conjugated secondary antibody] at 4 ℃ overnight. Images were acquired using the ImageQuant Las 4000 system (GE Healthcare, Massachusetts, USA), and the density of each protein band was calculated using Image J software.
Construction of the cell line-derived xenograft (CDX) model
Animal experiments were performed under a project license (No. LA2019173) granted by the Institutional Animal Care and Use Committee of the Peking University, in compliance with its guidelines for the care and use of animals. A protocol was prepared before the study without registration. Male athymic BALB/c nude mice (4–6 weeks old) were housed under broken barrier-specific pathogen-free conditions, with 3 mice per cage. Considering animal welfare and economics, 3 mice were used in each group to decrease the individual differences. Animals were randomly assigned (using a random number generator) into treatment and control groups and evaluated by individuals blinded to treatment regime received. In addition, all subsequent operations were performed randomly to minimize potential confounders. All mice were maintained on a regular diurnal lighting cycle (12:12 light: dark) with ad libitum access to food (7012 Harlan Teklad LM-485 Mouse/Rat Sterilizable Diet) and water, with chopped corn cob as bedding. The CDX model was established by injecting siRNA-treated tumor cells (1×106 cells per mouse) into the bottom of the mouth. Considering the complex microenvironment in animals and the degradability of siRNA, ML264 (Selleck, Shanghai, China) was used to inhibit KLF5 expression in vivo. It was injected around the tumors every 2 days at 25 mg/kg and the vehicle solution was used as the negative control. Considering the oversized tumor might obstruct mice’s breathing, all mice were euthanized after 3 weeks and the tumors were excised for further analysis.
Immunofluorescence analysis
Tumor tissues were fixed overnight with 4% paraformaldehyde, dehydrated with a 30% sucrose aqueous solution, embedded in OCT reagent (Sakura, Tokyo, Japan), and sliced to a thickness of 10 µm (Leica, Wetzlar, Germany). The sections were incubated with primary antibodies against KLF5 (1:100), BIRC5 (1:500), and HRP-conjugated secondary antibodies (1:2,000). Apoptosis of tumor tissues was assessed using a TUNEL kit (Solarbio). Representative photomicrographs were obtained using an Eclipse 90i microscope (Nikon, Tokyo, Japan).
Statistical analysis
All statistical analyses were performed using SPSS (version 27.0) and R Studio (version 4.0.2). All data are presented as the mean ± standard deviation of at least 3 independent experiments. The Student’s t-test was used to evaluate differences between the 2 groups. Statistically significant data were presented, and P<0.05 was set as the threshold for statistical significance.
Results
Functional enrichment analysis of KLF5
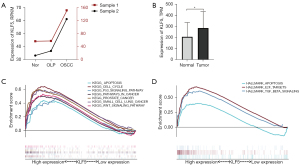
As shown in Figure 1A, the results demonstrated that KLF5 expression increased from normal tissues to OLP and then to OSCC tissues in the 2 clinical samples. In addition, significant differential expression of KLF5 between normal and OSCC tissues was observed in the TCGA dataset, with higher expression in OSCC (Figure 1B). Further analysis suggested that the high-expression group was mainly enriched in apoptosis, the cell cycle, the P53 signaling pathway, pathways in cancer, and the wingless/integrated (Wnt) signaling pathway in the KEGG pathways (Figure 1C), while apoptosis, E2F targets, and transforming growth factor-β (TGF-β) pathway were enriched in hallmarks (Figure 1D).
KLF5 inhibited cell apoptosis in OPMDs and OSCC
Cell apoptosis was detected using annexin V/PI staining in DOK, CAL27, and SCC15 cells. As shown in Figure 2A-2D, the results indicated increased cell apoptosis after si-KLF5 treatment, suggesting that high expression of KLF5 could inhibit cell apoptosis.
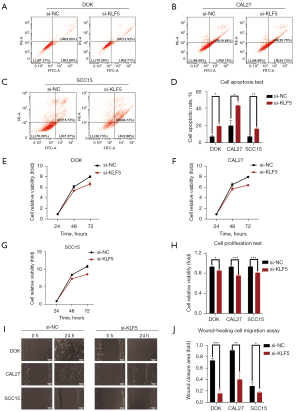
KLF5 promoted cell proliferation in OPMDs and OSCC
After siRNA-mediated knockdown of KLF5 in cells, we used CCK-8 assays to assess cell viability as a marker of proliferation 24, 48, and 72 h after transfection (Figure 2E-2G). We observed marked suppression of cell viability after KLF5 knockdown (Figure 2H).
KLF5 accelerated cell migration in OPMDs and OSCC
A wound-healing assay was used to evaluate the impact of KLF5 on cell migration (Figure 2I). DOK, CAL27, and SCC15 cells exhibited significantly reduced wound-healing ability after siRNA-mediated KLF5 knockdown (Figure 2J).
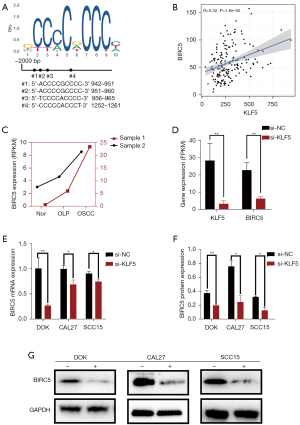
Low expression of KLF5 inhibited BIRC5 in vitro
As shown in Figure 3A, further analysis of the NCBI and JASPAR data showed 4 binding sites on the BIRC5 promoter region, even though we set the relative profile score threshold at 95%. The higher the threshold value, the more likely it is that the KLF5 transcription factor could combine with the BIRC5 promoter region. Further analysis of the TCGA dataset indicated that the expression levels of KLF5 and BIRC5 were positively correlated (Figure 3B). BIRC5 differential expression was confirmed in paired samples from the 2 patients, with the highest expression in OSCC tissues (Figure 3C). The analysis of the transcriptome sequencing data suggested that the expression of BIRC5 also decreased when we knocked down KLF5 expression in CAL27 cells (Figure 3D). Furthermore, qRT-PCR and western blot analysis showed lower expression of BIRC5 mRNA and protein after si-KLF5 treatment in OSCC and OPMD cells (Figure 3E-3G). Thus, KLF5 knockdown decreased BIRC5 expression in vitro.
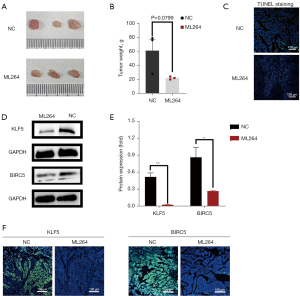
Animal experiments
CAL27 cells have been used to build animal models and contribute to the formation of OSCC tumors. After euthanasia, the tumors were removed from the nude mice and imaged (Figure 4A). As shown in Figure 4B, the tumor weight in the ML264 group was lower than that in the negative control group (P=0.0799). The results of the TUNEL assay showed a higher apoptosis rate in tumor tissues after ML264 treatment than in the control group (Figure 4C). Western blotting and immunohistochemical imaging of tumors dissected from nude mice showed that BIRC5 protein expression was decreased after ML264-induced inhibition of KLF5 in vivo (Figure 4D,4E). As shown in Figure 4F, the immunofluorescence signals for KLF5 and BIRC5 were darker after treatment with ML264. These results indicate that high KLF5 expression accelerates tumor growth, inhibits apoptosis, and affects BIRC5 expression in vivo.
Discussion
The current strategy for OSCC treatment is surgical excision combined with radiotherapy and chemotherapy, which not only causes large tissue defects but also damages the psychological health of the patients (12). In recent decades, targeted therapy has attracted increasing attention, and numerous potential targets have been explored for cancer therapy at the molecular level.
NGS is characterized by sequencing millions of molecules in parallel, making it possible to analyze the entire transcriptome and genome of a species in detail (13). It significantly decreases the cost and time, and it permits the quick and relatively cheap sequencing of multiple loci in the genome. Meanwhile, it minimizes the number of fragment cloning during gene hunting studies (14). Potential targets can be predicted by exploring differentially expressed genes using bioinformatics analysis. In addition, it is possible to identify potential molecular mechanisms and related pathways using enrichment clustering analysis of sequencing data (15).
There are many clinicopathological factors to influence the prognosis of patients with OSCC, and Tumor Node Metastasis (TNM) staging system is internationally accepted (16). Some of KLFs are identified as critical molecules in OSCC by involving in above factors. In OSCC cells, high-expression KLF16 (17) and low-expression KLF2 (18) markedly promote cell malignant behavior, including cell proliferation, apoptosis, migration, invasion, and epithelial-mesenchymal transition (EMT). In clinical samples, KLF4 (19), KLF7 (20) and KLF8 (21) overexpression accelerate OSCC invasiveness, lymph node metastasis or distant metastasis. In addition, KLF4 is highly expressed in the cut margins in recurrent OSCC, indicating it serves as a marker to predict recurrence (22). KLF5 functions as a cancer promoter in other tumors and has been reported to affect cell stemness, proliferation, apoptosis, autophagy, and migration (23). KLF5 is markedly upregulated in gastric cancer and its expression is positively correlated to TNM staging (24). KLF5 promotes TNFα-induced apoptosis and facilitates the initiation and progression of prostate cancer (25). KLF5 has also been identified as an oncogene in breast cancer and hepatocellular carcinoma, and its expression is directly associated with a poor prognosis (26,27). Interestingly, there remain a few controversies regarding KLF5 function in esophageal squamous cell carcinoma (28). However, KLF5 has emerged as an oncogene that promotes OSCC occurrence and development.
The accuracy of NGS is associated with tissue availability, handling and processing, nucleic acid yield, and quality may considerably influence the results (29). In our previous research, NGS technology and further analysis were applied to paired tumorous and premalignant tissues and adjacent normal mucosa from 2 patients diagnosed with OSCC and OLP to reduce the individual differences and all samples were treated in same condition. The results showed that KLF5 expression gradually increased from normal mucosa tissue to OLP and then to OSCC tissue. We found that the expression of KLF5 in OSCC was also significantly increased in the TCGA data. GSEA showed that it was closely related to the P53, E2F, and TGF-β signaling pathways, and all of them were closely related to the occurrence and development of tumors as central molecules. P53 and E2F are identified as tumor suppressors, whereas the dual function of TGF-β is also strongly associated with the development of various malignant tumors, including OSCC (30-32). In OSCC, TP53 mutations and high expression of E2F7 play carcinogenic roles and indicate worse survival and resistance to chemotherapy and radiotherapy (33,34). Analysis of clinical samples suggests that patients with high TGF-β expression in the tumor center have a higher recurrence rate and worse extent of invasion (35). Our bioinformatics analysis showed that high KLF5 expression was closely related to the initiation and progression of OSCC. Further results demonstrated that it could accelerate carcinogenesis by promoting cell proliferation and migration, while inhibiting apoptosis. ML264 inhibits KLF5 expression by directly targeting its transcription in colorectal cancer (36). Animal experiments showed that the tumor weight decreased after inhibiting KLF5 expression with ML264 in athymic BALB/c nude mice.
To further explore the oncogenic mechanism of KLF5, we found that BIRC5 expression also gradually increased from normal mucosa tissues to OLP and then to OSCC tissues. Its expression was positively correlated with KLF5 in clinical samples and the TCGA dataset. In previous studies, BIRC5 is a tumor-specific gene involved in cell apoptosis, accelerating tumor-associated angiogenesis, and resisting anticancer therapies (37,38). BIRC5 expression is higher in prostate cancer and its high expression is associated with greater tumor aggressiveness and progression (39). High expression of BIRC5 protein also indicates lower 5-year survival rates in patients with colorectal cancer (40).
Further exploration was performed to reveal the potential relationship between KLF5 and BIRC5. We searched the promoter sequence of BIRC5 on NCBI (https://www.ncbi.nlm.nih.gov/) and further analyzed whether there was a binding site for the KLF5 transcription factor in the promoter region of BIRC5 in the JASPAR database (https://jaspar.genereg.net/). We predicted 4 binding sites in which the KLF5 transcription factor could combine with the BIRC5 promoter region at a 95% relative profile score threshold. The results of qRT-PCR and western blotting showed that BIRC5 expression decreased after KLF5 inhibition in vitro and in vivo.
Overall, our results indicated that KLF5 expression has increased in OSCC, and its high expression could inhibit cell apoptosis while promoting cell proliferation and migration. KLF5 may be a potential diagnostic and therapeutic target in OSCC, likely by regulating BIRC5. However, further research is needed to explore the detailed mechanism by which KLF5 regulates BIRC5 expression.
Acknowledgments
Funding: This study was supported by a grant from the National Natural Science Foundation of China (No. 81902766).
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3728/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3728/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3728/coif). All authors report that this study was supported by a grant from the National Natural Science Foundation of China. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Peking University Institutional Review Board (approval No. IRB00001052–12037) and informed consent was taken from both patients. Animal experiments were performed under a project license (No. LA2019173) granted by the Institutional Animal Care and Use Committee of the Peking University, in compliance with its guidelines for the care and use of animals.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chai AWY, Lim KP, Cheong SC. Translational genomics and recent advances in oral squamous cell carcinoma. Semin Cancer Biol 2020;61:71-83. [Crossref] [PubMed]
- Diebold S, Overbeck M. Soft Tissue Disorders of the Mouth. Emerg Med Clin North Am 2019;37:55-68. [Crossref] [PubMed]
- Warnakulasuriya S, Ariyawardana A. Malignant transformation of oral leukoplakia: a systematic review of observational studies. J Oral Pathol Med 2016;45:155-66. [Crossref] [PubMed]
- Wang YY, Xiao LY, Wu PC, et al. Orabase-formulated gentian violet effectively improved oral potentially malignant disorder in vitro and in vivo. Biochem Pharmacol 2020;171:113713. [Crossref] [PubMed]
- Walsh T, Warnakulasuriya S, Lingen MW, et al. Clinical assessment for the detection of oral cavity cancer and potentially malignant disorders in apparently healthy adults. Cochrane Database Syst Rev 2021;12:CD010173. [PubMed]
- McConnell BB, Kim SS, Yu K, et al. Krüppel-like factor 5 is important for maintenance of crypt architecture and barrier function in mouse intestine. Gastroenterology 2011;141:1302-13, 1313.e1-6.
- Zhu N, Gu L, Findley HW, et al. KLF5 Interacts with p53 in regulating survivin expression in acute lymphoblastic leukemia. J Biol Chem 2006;281:14711-8. [Crossref] [PubMed]
- Dong Z, Yang L, Lai D. KLF5 strengthens drug resistance of ovarian cancer stem-like cells by regulating survivin expression. Cell Prolif 2013;46:425-35. [Crossref] [PubMed]
- Li F, Aljahdali I, Ling X. Cancer therapeutics using survivin BIRC5 as a target: what can we do after over two decades of study? J Exp Clin Cancer Res 2019;38:368. [Crossref] [PubMed]
- Troiano G, Guida A, Aquino G, et al. Integrative Histologic and Bioinformatics Analysis of BIRC5/Survivin Expression in Oral Squamous Cell Carcinoma. Int J Mol Sci 2018;19:2664. [Crossref] [PubMed]
- Shi W, Yang J, Li S, et al. Potential involvement of miR-375 in the premalignant progression of oral squamous cell carcinoma mediated via transcription factor KLF5. Oncotarget 2015;6:40172-85. [Crossref] [PubMed]
- Bossi P, Alfieri S, Strojan P, et al. Prognostic and predictive factors in recurrent and/or metastatic head and neck squamous cell carcinoma: A review of the literature. Crit Rev Oncol Hematol 2019;137:84-91. [Crossref] [PubMed]
- Hess JF, Kohl TA, Kotrová M, et al. Library preparation for next generation sequencing: A review of automation strategies. Biotechnol Adv 2020;41:107537. [Crossref] [PubMed]
- Pipis M, Rossor AM, Laura M, et al. Next-generation sequencing in Charcot-Marie-Tooth disease: opportunities and challenges. Nat Rev Neurol 2019;15:644-56. [Crossref] [PubMed]
- Gong L, Zhang D, Dong Y, et al. Integrated Bioinformatics Analysis for Identificating the Therapeutic Targets of Aspirin in Small Cell Lung Cancer. J Biomed Inform 2018;88:20-8. [Crossref] [PubMed]
- Chi AC, Day TA, Neville BW. Oral cavity and oropharyngeal squamous cell carcinoma--an update. CA Cancer J Clin 2015;65:401-21. [Crossref] [PubMed]
- Qin SY, Li B, Chen M, et al. MiR-32-5p promoted epithelial-to-mesenchymal transition of oral squamous cell carcinoma cells via regulating the KLF2/CXCR4 pathway. Kaohsiung J Med Sci 2022;38:120-8. [Crossref] [PubMed]
- Yang L, Shi YL, Ma Y, et al. Silencing KLF16 inhibits oral squamous cell carcinoma cell proliferation by arresting the cell cycle and inducing apoptosis. APMIS 2022;130:43-52. [Crossref] [PubMed]
- Yoshihama R, Yamaguchi K, Imajyo I, et al. Expression levels of SOX2, KLF4 and brachyury transcription factors are associated with metastasis and poor prognosis in oral squamous cell carcinoma. Oncol Lett 2016;11:1435-46. [Crossref] [PubMed]
- Ding X, Wang X, Gong Y, et al. KLF7 overexpression in human oral squamous cell carcinoma promotes migration and epithelial-mesenchymal transition. Oncol Lett 2017;13:2281-9. [Crossref] [PubMed]
- Li X, Ma C, Zhang L, et al. LncRNAAC132217.4, a KLF8-regulated long non-coding RNA, facilitates oral squamous cell carcinoma metastasis by upregulating IGF2 expression. Cancer Lett 2017;407:45-56. [Crossref] [PubMed]
- Roy S, Kar M, Roy S, et al. KLF4 expression in the surgical cut margin is associated with disease relapse of oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol 2019;128:154-65. [Crossref] [PubMed]
- Wang X, Qiu T, Wu Y, et al. Arginine methyltransferase PRMT5 methylates and stabilizes KLF5 via decreasing its phosphorylation and ubiquitination to promote basal-like breast cancer. Cell Death Differ 2021;28:2931-45. [Crossref] [PubMed]
- Chen P, Qian XK, Zhang YF, et al. KLF5 promotes proliferation in gastric cancer via regulating p21 and CDK4. Eur Rev Med Pharmacol Sci 2020;24:4224-31. [PubMed]
- Shi Q, Gao Y, Xu S, et al. Krüppel-like factor 5 promotes apoptosis triggered by tumor necrosis factor α in LNCaP prostate cancer cells via up-regulation of mitogen-activated protein kinase kinase 7. Urol Oncol 2016;34:58.e11-8. [Crossref] [PubMed]
- Tong D, Czerwenka K, Heinze G, et al. Expression of KLF5 is a prognostic factor for disease-free survival and overall survival in patients with breast cancer. Clin Cancer Res 2006;12:2442-8. [Crossref] [PubMed]
- An T, Dong T, Zhou H, et al. The transcription factor Krüppel-like factor 5 promotes cell growth and metastasis via activating PI3K/AKT/Snail signaling in hepatocellular carcinoma. Biochem Biophys Res Commun 2019;508:159-68. [Crossref] [PubMed]
- Luo Y, Chen C. The roles and regulation of the KLF5 transcription factor in cancers. Cancer Sci 2021;112:2097-117. [Crossref] [PubMed]
- Chen H, Luthra R, Goswami RS, et al. Analysis of Pre-Analytic Factors Affecting the Success of Clinical Next-Generation Sequencing of Solid Organ Malignancies. Cancers (Basel) 2015;7:1699-715. [Crossref] [PubMed]
- Kotler E, Shani O, Goldfeld G, et al. A Systematic p53 Mutation Library Links Differential Functional Impact to Cancer Mutation Pattern and Evolutionary Conservation. Mol Cell 2018;71:178-190.e8. [Crossref] [PubMed]
- Kent LN, Leone G. The broken cycle: E2F dysfunction in cancer. Nat Rev Cancer 2019;19:326-38. [Crossref] [PubMed]
- Colak S, Ten Dijke P. Targeting TGF-β Signaling in Cancer. Trends Cancer 2017;3:56-71. [Crossref] [PubMed]
- Lindemann A, Takahashi H, Patel AA, et al. Targeting the DNA Damage Response in OSCC with TP53 Mutations. J Dent Res 2018;97:635-44. [Crossref] [PubMed]
- Zhou P, Xiao L, Xu X. Identification of E2F transcription factor 7 as a novel potential biomarker for oral squamous cell carcinoma. Head Face Med 2021;17:7. [Crossref] [PubMed]
- Lu Z, Ding L, Ding H, et al. Tumor cell-derived TGF-β at tumor center independently predicts recurrence and poor survival in oral squamous cell carcinoma. J Oral Pathol Med 2019;48:696-704. [Crossref] [PubMed]
- Ruiz de Sabando A, Wang C, He Y, et al. ML264, A Novel Small-Molecule Compound That Potently Inhibits Growth of Colorectal Cancer. Mol Cancer Ther 2016;15:72-83. [Crossref] [PubMed]
- Velculescu VE, Madden SL, Zhang L, et al. Analysis of human transcriptomes. Nat Genet 1999;23:387-8. [Crossref] [PubMed]
- Lin TY, Chan HH, Chen SH, et al. BIRC5/Survivin is a novel ATG12-ATG5 conjugate interactor and an autophagy-induced DNA damage suppressor in human cancer and mouse embryonic fibroblast cells. Autophagy 2020;16:1296-313. [Crossref] [PubMed]
- Hennigs JK, Minner S, Tennstedt P, et al. Subcellular Compartmentalization of Survivin is Associated with Biological Aggressiveness and Prognosis in Prostate Cancer. Sci Rep 2020;10:3250. [Crossref] [PubMed]
- Or CR, Huang CW, Chang CC, et al. Obatoclax, a Pan-BCL-2 Inhibitor, Downregulates Survivin to Induce Apoptosis in Human Colorectal Carcinoma Cells Via Suppressing WNT/β-catenin Signaling. Int J Mol Sci 2020;21:1773. [Crossref] [PubMed]
(English Language Editor: C. Betlazar-Maseh)


