Mechanisms of immune-related differentially expressed genes in thyroid-associated ophthalmopathy based on the GEO database
Introduction
Thyroid-associated ophthalmopathy (TAO), also known as Graves’ disease (GD), is an autoimmune disease that is rarely seen in patients with chronic autoimmune thyroiditis with normal or hypothyroidism, and is characterized by infiltrative lesions in the posterior and periorbital tissues of the eye (1). TAO most commonly occurs in women aged 40–60 years, and it has an overall prevalence of approximately 0.0422 per 1,000 people; however, studies have reported a wide variation in the prevalence of TAO in patients with different types of GD (2-4). Further, 20–50% of patients with GD have ocular involvement, and some patients disease progression, leading to dysthyroid optic neuropathy (DON), exposed corneal ulcers, etc. (5,6). The pathogenesis of TAO is complex, and it is currently believed that it is mainly related to environmental, genetic, and immune factors (7). Notably, immune factors are often considered the core factors involved in the occurrence and progression of TAO, and the pathogenesis of TAO can be studied from the perspective of immunity.
Many of the signs and symptoms of TAO are caused by an increase in pressure within the bony volume of the orbit due to the expansion of soft tissues within the orbit, pathomechanism mainly involving adipogenesis, glycosaminoglycan accumulation and inflammation (8). Previous studies have shown that orbital fibroblasts are the main effector cells of the TAO autoimmune response (9), and have a heterogeneous phenotype and function (8). Activated fibroblasts secrete a variety of cytokines that lead to uncontrolled immune responses (7,10). Several pathogenetic mechanisms of TAO have been proposed; however, a clear pathogenesis has not yet been fully elucidated.
Ribonucleic acid–sequencing (RNA-seq) is a 2nd-generation high-throughput sequencing technology that sequences RNAs from specific tissues or cells into complementary deoxyribonucleic acid (DNA) for quantitative and qualitative studies of gene expression, which can be used to explore the molecular mechanisms of disease and provide a scientific basis for the prevention and treatment of human diseases. This study sought to analyze the transcriptome information of TAO to further elucidate the immune mechanisms associated with TAO and to explore potential biomarkers to provide a scientific basis for the prevention and treatment of TAO. We present the following article in accordance with the STREGA reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3470/rc).
Methods
Data collection
The Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) is a public genomics database. We searched the database using “thyroid-associated ophthalmopathy” as the keyword and included data sets with sample sizes ≥10. GSE105149 and GSE58331 were ultimately included in this analysis. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Immune-related genes
We downloaded the immune-related genes from the Immunology Database and Analysis Portal (ImmPort) (https://www.immport.org/home) database, which contains a total of 2,498 immune-related genes from 17 immune classifications.
Differential expression analysis
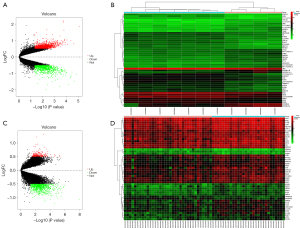
We extracted 2 data sets (i.e., GSE105149 and GSE58331) comprising the data of TAO and normal patients. The data were normalized using the limma package in R language, the differentially expressed genes (DEGs) were analyzed using the limma package, and the threshold of significance was set as follows: fold of variation >0.5 and a P value <0.5. The results were analyzed, the pheatmap package was used to construct a heat map to visualize the results, and the ggplot2 package was used to construct a volcano map to visualize the distribution of the DEGs.
Establishing core genes
Venny version 2.1 was used to obtain the core key genes of immune-related thyroid eye disease by inputting the DEGs and immune-related genes in two datasets GSE105149 and GSE58331, and the results were visualized by Veen plots.
GO and KEGG analysis
To determine the function of the target genes, we performed a gene ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses using the R package clusterProfiler (Papaemmanuil et al., 2016). The GO terminology comprised the following 3 components: biological processes (BPs), cell components (CCs), and molecular functions (MFs). A q value <0.05 was considered statistically significant.
Protein-protein interaction (PPI) and GGI
We used the immune-related key genes of TAO in the Search Tool for the Retrieval of Interacting Genes (STRING) database (https://string-db.org/) to construct a functional PPI network. Interactions with a composite score >0.4 were considered statistically significant. The immune-related TAO key genes were then entered into the online tool GeneMinia (http://genemania.org/), to construct the gene-gene interaction (GGI) network.
MiRNA target gene network prediction
In disease states, microRNAs (miRNAs) affect gene expression through post-transcriptional control. This study used the miRWalk database (http://miRWalk.umm.uni-heidelberg.de/) to search for the miRNAs associated with the DEGs. miRWalk is a publicly available comprehensive database of miRNA target genes that includes predicted and experimentally validated miRNA-target interaction pairs from humans, mice, rats, dogs, and bovine. We imported immune-related TAO key genes into the miRWalk database, set the score as 1, and validated with the miRTarBase database (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) as a screening condition, and imported the results into Cytoscape (version 3.9.0) to construct a regulatory network structure for the immune-related key genes in TAO.
Immune infiltration
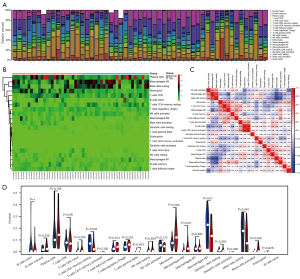
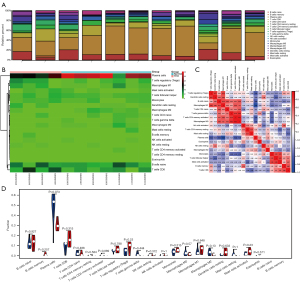
We used the CIBERSORT online website (http://CIBERSORT.stanford.edu/) to predict the proportion of 22 immune cells in all the sample data sets. We set 500 replicates, and samples with a P value <0.05 were considered statistically significant. We visualized the results using the ggplot2 package in R. The bar plot and heatmap reflect the relative content of the 22 immune cells in each sample. The correlation heatmap analyzed and visualized the correlation between the immune cells. The violin map revealed the differences in the content of the immune cells in different groups.
Immune correlation analysis
The matrix results for immune infiltration were combined with the normalized data set of the immune-related key genes of TAO and analyzed to determine the correlations. The results were visualized and analyzed using the ggcorrplot package in R language to construct bar plots to assess the correlations between the immune-related genes and the immune cells. Correlation coefficients of 0.4–0.7 were considered moderate, and correlation coefficients >0.7 were considered high.
Drug sensitivity analysis
Using the genecard database (https://www.genecards.org/), the TED therapeutic target genes were searched using the keyword, “thyroid-associated ophthalmopathy,” and then intersected with the immune-related TAO key genes to identify additional immune-related genes. The key genes of TAO were crossed with immune-related genes. Finally, using the Drug-Gene Interaction Database (DGIdb; https://www.dgidb.org/), potential drugs targeting the intersecting genes were predicted and visualized as network modules using Cytoscape, and the genes that could be used as therapeutic targets were identified as the immune-related key genes for TAO.
Receiver operator characteristic (ROC) curves and diagnostic model
ROC curves for the immune-related therapeutic genes in TAO were constructed using the “rms” package in R language to assess the diagnostic efficacy of the genes in both data sets, and the areas under the curve (AUCs) were used to assess the diagnostic efficacy of the genes; AUCs of 0.6–0.7 indicated low efficacy, AUCs of 0.7–0.8 indicated moderate efficacy, AUCs >0.8 indicated high efficacy. Subsequently, the diagnostic model for the diagnosis of TAO was constructed by the survminer package and the rms package, and the diagnostic efficacy of the model was assessed by the concordance index (C index), and the accuracy of the model was assessed by the correction curve.
Results
TAO immune-related differentially expressed genes (irDEGs)
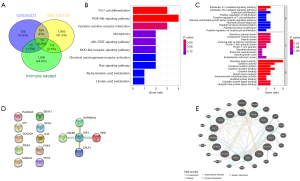
DEGs were identified for the GSE105149 and GSE58331 data sets, respectively, and the results showed that the DEGs were significantly more upregulated and downregulated in the GSE58331 data set than the GSE105149 data set (see Figure 1). There was a total of 200 DEGs in the GSE105149 and GSE58331 data sets, of which 15 genes were immune-related (see Figure 2A).
GO and KEGG enrichment analysis of irDEGs
Using R software, biological function and pathway enrichment analyses were performed based on the TAO irDEGs. The top 10 BPs, CCs, MFs, and 10 KEGG pathways were selected according to the number and significance of gene enrichment, and the bar and bubble plots were drawn separately. In relation to the GO BPs, the main pathways included the interleukin (IL)-27-mediated signaling pathway, IL-35-mediated signaling pathway, secretory granule lumen, cytoplasmic vesicle lumen, receptor ligand activity, cytokine activity, T helper 17 (Th17) cell differentiation, phosphatidylinositol-3-kinase and protein kinase B (PI3K-Akt) signaling pathway, cytokine-cytokine receptor interaction, Janus kinase and signal transducer and activator of transcription (JAK-STAT) signaling pathway, and other KEGG pathways (see Figure 2B,2C).
PPI and GGI network construction
The PPI and GGI analyses of the DEGs were performed using the STRING database and GeneMinia tool. In the PPI network, there were 5 nodes and 4 edges composed of Janus kinase 1 (JAK1), heat shock protein 90-α (HSP90AA1), colony stimulating factor 3 receptor (CSF3R), cytokine receptor-like factor 1 (CRLF1), and Epstein-Barr virus induced 3 (EBI3), and in the GGI analysis, 29 interacting genes were enriched (see Figure 2D,2E).
MicroRNA target gene network analysis
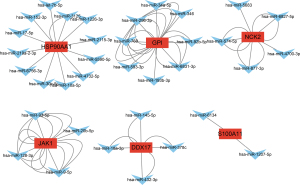
To further explore the potential miRNAs of these DEGs, we constructed an immune-related TAO regulatory network, which showed miRNAs for a total of 6 key genes [i.e., HSP90AA1, glucose-6-phosphate isomerase (GPI), NCK adaptor protein 2 (NCK2), JAK1, DDC17, and S100 calcium binding protein A11 (S100A11)] (see Figure 3).
Immune-infiltration analysis
To further explore the role of immune cells in TAO, we analyzed the GEO data sets separately. The results showed that cluster of differentiation (CD)4+ T cells, monocytes, M0 macrophages, and Mast cells were significantly elevated in TAO, while M2 macrophages were significantly reduced in TAO. In the immune cell correlation analysis, the CD4+ T cells and naïve B cells were significantly positively correlated and significantly negatively correlated with activated natural killer (NK) cells, respectively, while Mast cells were positively correlated with plasma cells and negatively correlated with M2 macrophages (see Figures 4,5).
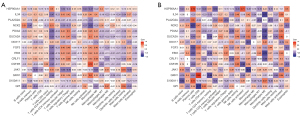
Immunological correlation analysis
A correlation analysis was performed between the irDEGs and the immune-infiltration results. In the GSE105149 data set, HSP90AA1, S100A11, and phospholipase A 2 group IIA (PLA2G2A) were positively correlated with plasma cells, Gastrokine 1 (GKN1) was positively correlated with CD8 T cells, and GPI was positively correlated with CD4 naïve T cells. Conversely, in the GSE58331 data set, CSF3R and GKN1 were positively correlated with monocytes, HSP90AA1, NCK2 and JAK1 were positively correlated with resting memory CD4 T cells (see Figure 6).
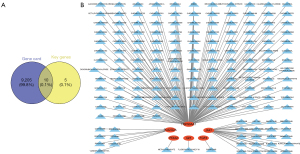
Drug sensitivity analysis
To explore the potential therapeutic targets of irDEGs, we performed a drug sensitivity prediction analysis. The results showed that there were 10 intersecting genes, among which JAK1, HSP90AA1, PLA2G2A, FGF3, GPI, and PDIA2 are six key genes, and more than 100 potential targeted drugs have been predicted (see Figure 7).
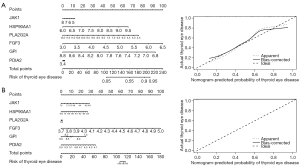
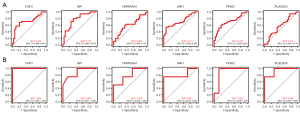
Construction of ROC curves and diagnostic models
To develop risk-prediction models, we first performed a diagnostic efficacy analysis of immune-related differential expression memory cells and finally included a total of 6 genes (i.e., JAK1, HSP90AA1, PLA2G2A, FGF3, GPI, and PDIA2) in the models (see Figure 8). The models were constructed for the 2 GEO datasets separately, and the results showed that the models all had good predictive efficacy (see Figure 9).
Discussion
TAO is an autoimmune disease with a complex etiology and poor treatment outcomes. The genetic and immune factors of TAO and anti-immune therapy are receiving increasing clinical attention and are being studied more and more. Previous studies on the mechanisms of TAO have focused on orbital adipose tissue and extraocular muscles, but little research has been conducted on the lacrimal gland (11-13). In this study, 15 irDEGs, represented by JAK1, HSP90AA1, PLA2G2A, FGF3, GPI, and PDIA2, were screened by a differential analysis of TAO genes and normal human lacrimal gland expressed genes, and the intersecting immune genes were identified.
JAK1 is a large, widely expressed membrane-associated phosphoprotein that activates IL-6 and subsequently induces epithelial mesenchymal transition, which promotes proliferative vitreoretinopathy (14). HSP90AA1 is a ubiquitously expressed molecular chaperone that is efficiently expressed in response to stimuli, such as infection, trauma, and tumors, is involved in tumor development, and plays important roles in cell cycle regulation, gene expression, DNA damage, and carcinogenesis (15). Due to the lack of reports on TAO, HSP90AA1 may be a priority for future research. Secretory PLA2G2A is a phospholipase that plays a role in atherogenesis, inflammation, and host defense by increasing the metabolic rate and increasing glucose use in response to thyroid hormones (16). GPI is a member of the glucose phosphate isomerase protein family that plays an important role in glucose metabolism and can be secreted extracellularly to function as a growth factor or cytokine (17,18). PDIA2 belongs to the larger redox thioredoxin gene family of PDI, and the PDIA2 protein is a glycoprotein located in the endoplasmic reticulum with a high affinity for estrogen, which can act as an intracellular estrogen regulator in vitro and in vivo and may play a role in regulating immunity and redox reaction (19-22). Furthermore, our findings suggest that these key genes are closely positively or negatively associated with immune cell enrichment and are involved in regulating the immune microenvironment of TAO. The PPI core genes screened in this study are involved in several biologically important behaviors that are consistent with the complex mechanisms of TAO, further indicating the reliability of the prediction.
Our enrichment functional analysis of the screened irDEGs suggest that the BPs of TAO are closely related to immune regulation. Among them, the GO enrichment analysis suggested that the immune genes were mainly involved in BPs, such as T cells, lymphocytes and leukocytes, and IL, which further indicates that TAO is an immune process involving multiple inflammatory cells and factors. Inflammatory cytokines are synthesized and secreted by immune and non-immune cells in response to stimulation and can regulate the immune response and cell growth and differentiation by binding to the corresponding receptors. Previous studies have confirmed that inflammatory cells and cytokines are closely associated to TAO and are informative in the diagnosis of TAO (23-25).
The KEGG analysis showed that DEGs are closely associated with Th17 cell differentiation, the PI3K-Akt signaling pathway, the cytokine-cytokine receptor interaction, and the JAK-STAT signaling pathway. Studies have shown that the IL-17 signaling pathway is associated with the severity of TAO, and IL-17 may be an indicator of TAO disease activity evaluation (26-28). The PI3K/AKT pathway is not only closely related to the degradation of the extracellular matrix, but also to the malignant transformation of cells, and its key role in the development and progression of thyroid cancer has been confirmed both in vitro and in vivo (29-31). The above multiple pathways suggest that the development of TAO disease and immunity are closely related, and the continuous activation of the immune system may promote the progression of TAO.
Currently, researches indicate that T cell-mediated immunity promotes the development of TAO by mechanisms mainly related to B cell activation, the promotion of adhesion molecule expression, and the production of inflammatory cytokines (32,33). CD4+ T cells can be divided into subsets of T helper cells, T follicular helper (Tfh) cells, and regulatory T cells (Tregs). In the peripheral blood of TAO and GD patients, the proportion of Th1 in TAO patients was significantly higher than that of GD patients, and the ratio of Th1/Th2 was positively correlated with the TAO disease activity score (32). In vitro cultures of orbital muscle and adipocytes in TAO showed that Th1 was the main cytokine secreted by these 2 cells (34). Previous studies have shown that the number of Tregs in the peripheral blood increases after rituximab treatment of TAO and that absolute Treg values may be a clinical predictor of TAO (35,36). Hu et al. found that the percentage of peripheral CD3+CD4−CD8− T cells was significantly lower in active TAO patients than inactive TAO patients and was negatively correlated with the activity of the disease course; thus, these may be potential markers (37). These studies focused on orbital tissue and peripheral blood and did not analyze lacrimal gland tissue. In our study, T cells, macrophages and mast cells were found to be significantly elevated in patients with TAO, and a positive correlation was found between T cells and B cells, and T cells negative correlation with NK cells. These relationships reflect the synergistic or antagonistic effects between different immune cells in the progression of TAO. The consistent elevation of T cells in extraocular muscles, peripheral blood and lacrimal glands strongly suggests that they are predictors of TAO and that targeting of T cells may be considered.
Glucocorticoids (GCs) have long been used in the treatment of various ocular diseases due to their powerful anti-inflammatory, anti-edema, and anti-neoangiogenic properties (38-40). In patients with TAO, a GC is the main treatment option, and the European group for GD orbitopathy recommends intravenous GC therapy as the 1st-line option for patients with moderately severe active TAO (41,42). According to the prognostic model we constructed, we can predict the prognosis of TAO patients, and for patients with JAK1, HSP90AA1, PLA2G2A, FGF3, GPI and PDIA2 mutations, we can combine the clinical experience of drug use and the predicted targets to select therapeutic drugs. However, the heavy use of GCs may be associated with a range of adverse events and may not be appropriate for patients with comorbidities.
In this study, we performed a drug sensitivity analysis of key genes for constructing the model and mining >100 potential target drugs to provide a basis for clinical treatment. However, this study had some limitations. The data in this study were obtained from RNA-seq results in the GEO database, and in-vivo and in-vitro experiments were not conducted. Thus, the prognostic assessment model still needs to be validated with a large number of multicenter samples. Our group will continue to work on this model in the future.
Conclusions
In summary, TAO is a thyroid organ-specific autoimmune disease, and the immunologic pathogenesis plays the most important role in its development. The inflammatory response also plays an important role in its pathogenesis. In this study, bioinformatics was used to analyze the BPs and immune infiltration of TAO lacrimal gland expression gene microarrays. Our findings can be used as a reference for subsequent research work.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3470/rc
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3470/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Smith TJ, Janssen JAMJL. Insulin-like Growth Factor-I Receptor and Thyroid-Associated Ophthalmopathy. Endocr Rev 2019;40:236-67. [Crossref] [PubMed]
- Bartley GB. The epidemiologic characteristics and clinical course of ophthalmopathy associated with autoimmune thyroid disease in Olmsted County, Minnesota. Trans Am Ophthalmol Soc 1994;92:477-588. [PubMed]
- Weiler DL. Thyroid eye disease: a review. Clin Exp Optom 2017;100:20-5. [Crossref] [PubMed]
- Neag EJ, Smith TJ. 2021 update on thyroid-associated ophthalmopathy. J Endocrinol Invest 2022;45:235-59. [Crossref] [PubMed]
- Chin YH, Ng CH, Lee MH, et al. Prevalence of thyroid eye disease in Graves' disease: A meta-analysis and systematic review. Clin Endocrinol (Oxf) 2020;93:363-74. [Crossref] [PubMed]
- Dolman PJ. Dysthyroid optic neuropathy: evaluation and management. J Endocrinol Invest 2021;44:421-9. [Crossref] [PubMed]
- Bahn RS. Current Insights into the Pathogenesis of Graves' Ophthalmopathy. Horm Metab Res 2015;47:773-8. [Crossref] [PubMed]
- Bahn RS. Graves' ophthalmopathy. N Engl J Med 2010;362:726-38. [Crossref] [PubMed]
- Wang ZM, Wang ZY, Lu Y. The role of cell mediated immunopathogenesis in thyroid-associated ophthalmopathy. Int J Ophthalmol 2019;12:1209-14. [Crossref] [PubMed]
- Smith TJ, Hegedüs L. Graves' Disease. N Engl J Med 2016;375:1552-65. [Crossref] [PubMed]
- Wu L, Liang Y, Song N, et al. Differential expression and alternative splicing of transcripts in orbital adipose/connective tissue of thyroid-associated ophthalmopathy. Exp Biol Med (Maywood) 2021;246:1990-2006. [Crossref] [PubMed]
- Wu L, Li L, Liang Y, et al. Identification of differentially expressed long non-coding RNAs and mRNAs in orbital adipose/connective tissue of thyroid-associated ophthalmopathy. Genomics 2021;113:440-9. [Crossref] [PubMed]
- Zhang YJ, Zheng LL, Zhu Y, et al. Differential expression and functional mechanism of TIMD4 gene in orbital adipose tissues of patients with thyroid-associated ophthalmopathy. J Biol Regul Homeost Agents 2021;35:197-202. [PubMed]
- Gahl WA, Vale AM, Pitot HC. Separation of putrescine oxidase and spermidine oxidase in foetal bovine serum with the aid of a specific radioactive assay of spermidine oxidase. Biochem J 1980;187:197-204. [Crossref] [PubMed]
- Condelli V, Crispo F, Pietrafesa M, et al. HSP90 Molecular Chaperones, Metabolic Rewiring, and Epigenetics: Impact on Tumor Progression and Perspective for Anticancer Therapy. Cells 2019;8:532. [Crossref] [PubMed]
- Kuefner MS, Deng X, Stephenson EJ, et al. Secretory phospholipase A2 group IIA enhances the metabolic rate and increases glucose utilization in response to thyroid hormone. FASEB J 2019;33:738-49. [Crossref] [PubMed]
- Kassahn D, Kolb C, Solomon S, et al. Few human autoimmune sera detect GPI. Nat Immunol 2002;3:411-2; author reply 412-3. [Crossref] [PubMed]
- Kroemer G, López-Otín C, Madeo F, et al. Carbotoxicity-Noxious Effects of Carbohydrates. Cell 2018;175:605-14. [Crossref] [PubMed]
- Desilva MG, Notkins AL, Lan MS. Molecular characterization of a pancreas-specific protein disulfide isomerase, PDIp. DNA Cell Biol 1997;16:269-74. [Crossref] [PubMed]
- Fu XM, Zhu BT. Human pancreas-specific protein disulfide isomerase homolog (PDIp) is an intracellular estrogen-binding protein that modulates estrogen levels and actions in target cells. J Steroid Biochem Mol Biol 2009;115:20-9. [Crossref] [PubMed]
- Galligan JJ, Petersen DR. The human protein disulfide isomerase gene family. Hum Genomics 2012;6:6. [Crossref] [PubMed]
- Turano C, Coppari S, Altieri F, et al. Proteins of the PDI family: unpredicted non-ER locations and functions. J Cell Physiol 2002;193:154-63. [Crossref] [PubMed]
- Kishazi E, Dor M, Eperon S, et al. Differential profiling of lacrimal cytokines in patients suffering from thyroid-associated orbitopathy. Sci Rep 2018;8:10792. [Crossref] [PubMed]
- Mandić JJ, Kozmar A, Kusačić-Kuna S, et al. The levels of 12 cytokines and growth factors in tears: hyperthyreosis vs euthyreosis. Graefes Arch Clin Exp Ophthalmol 2018;256:845-52. [Crossref] [PubMed]
- Song RH, Wang B, Yao QM, et al. Proteomics Screening of Differentially Expressed Cytokines in Tears of Patients with Graves' Ophthalmopathy. Endocr Metab Immune Disord Drug Targets 2020;20:87-95. [Crossref] [PubMed]
- Wei H, Guan M, Qin Y, et al. Circulating levels of miR-146a and IL-17 are significantly correlated with the clinical activity of Graves' ophthalmopathy. Endocr J 2014;61:1087-92. [Crossref] [PubMed]
- Shen J, Li Z, Li W, et al. Th1, Th2, and Th17 Cytokine Involvement in Thyroid Associated Ophthalmopathy. Dis Markers 2015;2015:609593. [Crossref] [PubMed]
- Fang S, Huang Y, Wang N, et al. Insights Into Local Orbital Immunity: Evidence for the Involvement of the Th17 Cell Pathway in Thyroid-Associated Ophthalmopathy. J Clin Endocrinol Metab 2019;104:1697-711. [Crossref] [PubMed]
- Yin X, Ren M, Jiang H, et al. Downregulated AEG-1 together with inhibited PI3K/Akt pathway is associated with reduced viability of motor neurons in an ALS model. Mol Cell Neurosci 2015;68:303-13. [Crossref] [PubMed]
- Xing M. Genetic alterations in the phosphatidylinositol-3 kinase/Akt pathway in thyroid cancer. Thyroid 2010;20:697-706. [Crossref] [PubMed]
- Hou P, Liu D, Shan Y, et al. Genetic alterations and their relationship in the phosphatidylinositol 3-kinase/Akt pathway in thyroid cancer. Clin Cancer Res 2007;13:1161-70. [Crossref] [PubMed]
- Xia N, Zhou S, Liang Y, et al. CD4+ T cells and the Th1/Th2 imbalance are implicated in the pathogenesis of Graves' ophthalmopathy. Int J Mol Med 2006;17:911-6. [Crossref] [PubMed]
- Huang Y, Fang S, Li D, et al. The involvement of T cell pathogenesis in thyroid-associated ophthalmopathy. Eye (Lond) 2019;33:176-82. [Crossref] [PubMed]
- Antonelli A, Ferrari SM, Corrado A, et al. Extra-ocular muscle cells from patients with Graves' ophthalmopathy secrete α (CXCL10) and β (CCL2) chemokines under the influence of cytokines that are modulated by PPARγ. Autoimmun Rev 2014;13:1160-6. [Crossref] [PubMed]
- Kahaly GJ, Shimony O, Gellman YN, et al. Regulatory T-cells in Graves' orbitopathy: baseline findings and immunomodulation by anti-T lymphocyte globulin. J Clin Endocrinol Metab 2011;96:422-9. [Crossref] [PubMed]
- Khanna D, Chong KK, Afifiyan NF, et al. Rituximab treatment of patients with severe, corticosteroid-resistant thyroid-associated ophthalmopathy. Ophthalmology 2010;117:133-139.e2. [Crossref] [PubMed]
- Hu H, Liang L, Ge Q, et al. Correlation between Peripheral T Cell Subsets and the Activity of Thyroid-Associated Ophthalmopathy. Int J Endocrinol 2022;2022:2705650. [Crossref] [PubMed]
- Bartalena L, Kahaly GJ, Baldeschi L, et al. The 2021 European Group on Graves' orbitopathy (EUGOGO) clinical practice guidelines for the medical management of Graves' orbitopathy. Eur J Endocrinol 2021;185:G43-67. [Crossref] [PubMed]
- Chen X, Abudukerimu B, Li Q, et al. Influence of 4-week or 12-week glucocorticoid treatment on metabolic changes in patients with active moderate-to-severe thyroid-associated ophthalmopathy. Clin Transl Sci 2021;14:1734-46. [Crossref] [PubMed]
- González-García A, Sales-Sanz M. Treatment of Graves' ophthalmopathy. Med Clin (Barc) 2021;156:180-6. [Crossref] [PubMed]
- Bartalena L, Baldeschi L, Boboridis K, et al. The 2016 European Thyroid Association/European Group on Graves' Orbitopathy Guidelines for the Management of Graves' Orbitopathy. Eur Thyroid J 2016;5:9-26. [Crossref] [PubMed]
- Hoang TD, Stocker DJ, Chou EL, et al. 2022 Update on Clinical Management of Graves Disease and Thyroid Eye Disease. Endocrinol Metab Clin North Am 2022;51:287-304. [Crossref] [PubMed]