
Modification of m6A mediates tissue immune microenvironment in calcific aortic valve disease
Introduction
Aortic valve calcification (AVC) is the most common cause of surgical aortic valve replacement across the globe (1,2). Individuals aged above 65 years are more likely to be affected by AVC (2–7% of the population) in developed countries (3), and it is also a frequent consequence of chronic kidney disease (4). The incidence of AVC has been growing over the last decade, a trend which is expected to continue in the future (5-7). The lack of effective conservative treatment of AVC means that surgery remains the only readily available choice (8-12).
In AVC, the valvular interstitial cells (VICs) undergo transdifferentiation and become osteoblast-like cells, leading to the mineralization of the tissue (8,13,14). A range of pathogenic stimuli, including increased phosphate (15,16), may promote the translocation of Runx2 (a transcription factor) to the nucleus, triggering the cell to change to an osteoblast phenotype. Pro-inflammatory cytokines, namely, tumor necrosis factor (TNF) and interleukin-1 (IL-1), are associated with AVC, and inflammation is a characteristic of AVC (17-19). Rajamannan et al. (2) at the National Heart, Lung, and Blood Institute have shed light on AVC through a variety of innovative methods. They discovered that AVC is a dynamic and intricate osteogenic process that is initiated and promoted by inflammation. As previously documented, abnormal hemodynamic forces experienced by valve leaflets, including shear stress, hypertension, or increased strain, may result in tissue remodeling and inflammation which can advance to calcification, stenosis, and heart valve failure (2,8). Furthermore, according to the findings of an immunohistochemistry investigation conducted on the heart valves of 285 AVC patients, 28.4% of the affected tissues showed considerable leukocyte infiltration and high TNF-expression due to chronic inflammation (18). In a mouse model with IL-1 receptor antagonist (IL-1Ra) deficiency, inflamed aortic valve stenosis was identified, and TNF played a role in the initiation of valvular calcification (20). However, more research is required to determine the potential mechanism of the advancement of AVC.
In eukaryotes, N6-methyladenosine (m6A) is a common and well-studied non-cap reversible marker found on messenger RNAs (mRNAs) and long non-coding RNAs (lncRNAs) (21,22). The methylation of m6A is a process in which many proteins are involved. These proteins may broadly be divided into 3 categories: writer complexes, m6A demethylases, and function executions (readers). Within these categories, WTAP, RBM15, RBM15B, METTL3, METTL14, METTL16, and KIAA1429 are among the writer complexes; FTO and ALKBH5, the 2 evaluated m6A demethylases, are among the erasers; HNRNPs, YTHDF1, YTHDF2, YTHDF3, YTHDC1, YTHDC2, YTHDC3, EIF3A, IGF2BP1, IGF2BP2, and IGF2BP3 are among the function executions (readers) (23-26). Recent studies have shown that m6A is required for a wide range of biological activities, including viral infection (27,28), stress (29), heat shock (30), and DNA damage (31). The m6A modification has an impact on molecular functions (MFs) such as RNA-protein interaction (32), RNA stability (33), and translation efficiency (34).
Moreover, m6A dysregulation is linked to many diseases, including various forms of cancer, such as leukemia and glioma (35,36). Increasing evidence suggests that dysregulated m6A regulators are intimately linked to various heart-related complications, including cardiac failure, cardiac hypertrophy, vascular calcium deposition, and pulmonary hypertension (37,38). A recent study conducted by Zhou et al. (14) showed that METTL3 enhances osteogenic differentiation in human aortic VICs via an m6A YTHDF2-dependent cascade. The findings above revealed a unique method for the modification of the m6A genes in AVC; nevertheless, the overall expression characteristics of all m6A regulators in AVC, as well as the downstream and upstream molecular processes for important m6A regulators, remain unknown.
According to recent findings, m6A modification is related to immune infiltration (associated with diseases) and inflammation. Meanwhile, research has shown a link between m6A modification-related diseases and aberrant immune regulation (39,40), where upregulated METTL3 stimulated dendritic cells (DCs) activation and maturation, whereas downregulated METTL3 suppressed T cell activation and aggregation. In addition, Han et al. (41) found that suppressing YTHDF1 increased CD8+ T cell tumor invasion and immunotherapy efficacy in a mouse tumor model. The exact mechanism by which AVC-related m6A modification leads to immune cell infiltration is unknown. To better understand immune regulation in AVC, a detailed investigation of the diverse immune infiltration patterns regulated by the m6A regulators is required. These patterns of m6A modification were found via the analysis of genomic data from AVC samples in this study. On the basis of these findings, we further investigated the association between the m6A modification pattern and immune infiltration in AVC, as well as the roles and underlying mechanisms of m6A modification patterns in the immune microenvironment (IME) in AVC. We found that m6A modification patterns were associated with diseases as well as immune cell infiltration. By investigating differentially expressed genes (DEGs) in distinct m6A modification patterns, we showed that the m6A phenotype-related hub genes influence disease features and immune cell infiltration characteristics in AVC patients. In addition, we found that the m6A phenotype-related hub genes also have high diagnostic efficiency for AVC patients. A heat map was constructed to depict the pathways in which different gene clusters might contribute. Additionally, because AVC-induced stenosis is typically only treatable surgically, we sought potential therapeutic targets for early intervention. We present the following article in accordance with the STREGA reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3627/rc).
Methods
Calcific aortic valve disease data set source and preprocessing
The Gene Expression Omnibus (GEO) database was queried for public data and complete clinical annotation. We collected the GSE102249 (42) (240 diseased samples), GSE51472 (43) (5 normal samples and 10 AVC samples), and GSE12644 (44) (10 normal samples and 10 AVC samples) datasets for further investigation (Table S1). For the GSE51472 dataset, 5 sclerotic aortic valve samples were removed because the possible other lesions of the sclerotic aortic valve affected the analysis results. To begin, we used the platform’s annotation file to perform gene name conversion for probe IDs separately. For multiple probes corresponding to the same gene, we used the average expression of the gene. Each data was normalized for subsequent analysis. Since they shared a similar platform and might be used to merge data, GSE51472 and GSE12644 were integrated into a merged dataset. The batch effect was also eliminated using the combat function of the ‘SVA’ package. We observed the results of batch effect removal using principal component analysis (PCA). A total of 21,655 target gene expression profiles were generated. Next, 23 m6A methylase regulators (i.e., writers, erasers, and readers) in 3 types were used with reference to a previous study (45) and use these 23 m6A methylase regulators for follow-up analysis (Table S2). Finally, we found that 19 m6A-related genes are expressed in samples of merged data set.
Differential expression of m6A-related genes and immune cells
Based on the Wilcoxon test, we further analyzed the differences of 19 m6A methylase regulators and 6 immune cells (IL-10, IL-11, IL-6, IL-15, IL-16, TNF) between normal and AVC samples. A P value <0.05 was considered a significant difference.
Immune cell assessment and differential expression
The combined data set was calculated based on the immune cell content of Cell-type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT) samples, and the difference in immune cell content between normal and AVC samples was assessed by the Wilcoxon test.
AVC samples of GSE102249 dataset classification based on 23 m6A methylation regulators
Based on 23 m6A methylation regulators, we found that 20 m6A-related genes are expressed in samples of GSE102249 data set. We performed Consensus Clustering analysis on the 240 samples of the GSE102249 dataset, and divided them into m6A clusters A and B. The optimal k value was determined by evaluating the point of inflection for the sum of the squared errors. The decline slowed down after k=i, and k=i was selected. We used PCA to quantify the effect of typing.
Generation of the gene co-expression network and evaluation of the vital modules
To identify DEGs among m6A cluster patterns, 8,464 genes and their associated gene expression profiles were identified among the m6A cluster patterns. The DEGs were defined as those with an adjusted P<0.05 and |log2fold change (FC)| ≥1.
The “WGCNA” R-package was utilized to evaluate the top 3,000 DEGs according to the P value and modules with a high degree of connectivity. Each module was constructed by evaluating the soft threshold power β for all components. Following the establishment of the soft threshold power value, the weighted gene co-expression network analysis (WGCNA) algorithm was utilized to build the co-expression module. The information on adjacent genes was converted into topological overlap, and the network connection of the genes was determined. Based on the dissimilarity of the topological overlap matrix (TOM), hierarchical clustering was carried out to categorize genes with identical expression profiles into modules (the minimum size of gene dendrogram =20; merged highly similar modules with height =0.85). The association between clinical characteristics and module eigengenes (MEs) was utilized to identify the modules that were related to the clinical traits. The relationship between gene expression profiles and the MEs was utilized to calculate the quantitative measurement of module membership in this study. It was determined that genes were significant based on the absolute value of the connection between genes and clinical traits. Then, relevant modules with an elevated level of value for clinical characteristics were evaluated (Table S3).
Functional enrichment analysis of genes in the key modules
The R package ‘clusterProfiler’ was employed to evaluate the functional enrichment analysis on key module genes. The findings of the Gene Ontology (GO) including MF, cellular component (CC), and biological function (BF) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were performed. A P value <0.05 was considered statistically significant.
Identification of the key genes in the key module
To identify the interacting genes, we entered genes from important modules into the Search Tool for the Retrieval of Interacting Genes/Proteins [STRING (3); score >0.4] that predicted the protein-protein interaction network (PPIN) (Table S4). Important genes were determined as those having a significance greater than 0.3 and module membership greater than 0.6 in the modules. Finally, the hub genes of the disease-associated module and those of the PPIN were intersected, generating 7 disease-associated key genes.
AVC samples of GSE102249 dataset classification based on 7 key genes
Based on 7 key genes, we performed Consensus Clustering analysis on the 240 samples of the GSE102249 dataset again and divided them into gene cluster-1 and 2. The optimal k value was determined by evaluating the point of inflection for the sum of the squared errors. The decline slowed down after k=i, and k=i was selected. We used PCA to quantify the effect of typing.
Immune cell assessment and function enrichment
The combined data set was calculated based on the immune cell content of CIBERSORT samples, and the difference in immune cell content between 2 subgroups was assessed by the Wilcoxon test. The R package ‘clusterProfiler’ was employed to evaluate the functional enrichment analysis on key module genes. We performed GO findings on gene cluster-1 and 2, including MF, CC, and BP, and KEGG analysis, respectively, and created a thermal figure display. A P value <0.05 was considered statistically significant.
Statistical analysis
The R software (The R Foundation for Statistical Computing, Vienna, Austria) was employed to evaluate statistical analyses. The P values (bilateral) <0.05 were considered statistically significant.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
The landscape of m6A regulators and the tissue IME in AVC
Our study used datasets from the GEO (GSE51472 and GSE12644, Table S1). We used PCA analysis to observe the combined dataset batch effects after removing the combined results (Figure S1). We identified 23 m6A regulators based on previously published data (45), including 5 “writers”, 2 “erasers”, and 12 “readers” (Table S2). However, the gene profile of the combined dataset contained only 19 m6A methylation regulators. Figure 1A illustrates the positions of m6A regulators on the chromosomes. To identify the impact of the m6A regulators on the development of AVC, the transcriptional level of m6A regulators in AVC and healthy tissues was determined. The data indicated that the expression of “readers” (i.e., FMR1, HNRNPA2B1, HNRNPC, LRPPRCYTHDC1, YTHDC2, and YTHDF1) was significantly lower in the AVC tissues than in the normal tissues (P<0.01) (Figure 1B,1C). The expression of “writers” (i.e., METTL14) and “erasers” (i.e., FTO) was also significantly lower in the AVC tissues than in the normal tissues (P<0.01) (Figure 1B,1C). The underlined findings indicated that m6A regulators showed a high degree of heterogeneity in their expressional landscapes between normal and AVC patients, implying that differential m6A regulator expression might influence disease progression. Furthermore, we evaluated the characteristics of immune cell infiltration in AVC tissues and identified inflammatory factor IL-10 (P<0.05) (Figure 1D,1E) and M0 and M2 macrophages (P<0.05) (Figure 1F,1G). Compared to normal tissues, AVC tissues were enriched in IL-10 and M0 macrophages, which indicated an upregulation of the inflammatory factor. Conversely, M2 macrophages were more abundant in normal tissues, implying that infiltration of M2 macrophages may inhibit the progression of calcification. These findings suggested that changes in the aortic valve tissue’s IME might be the predominant pathogenic factor for AVC, and the infiltration of local M0 macrophages and the inflammatory factor IL-10 might promote AVC progression.
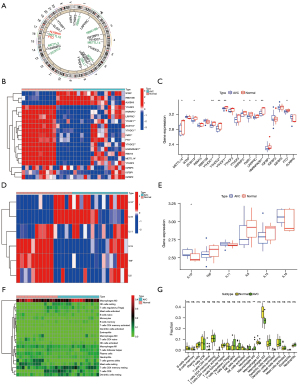
The m6A methylation modification patterns are facilitated by 19 m6A regulators in AVC patients
The m6A modification patterns were characterized using the R package ‘ConsensusClusterPlus’ based on the 20 m6A regulators. The fraction of unclear clustering assessment and the consistency of m6A regulator expression were used to determine the optimal clustering stability (k=2–10), and k=2 was obtained (Figure 2A,2B). We categorized the modification patterns of 240 AVC samples in GSE102249 using unsupervised clustering and identified 23 m6A regulators, including 5 “writers”, 2 “erasers”, and 9 “readers” (Table S2). Finally, 2 varied modification patterns were found, namely, 124 cases in cluster A and 116 cases in cluster B (Table S5). The expression of these m6A regulators could be used to differentiate the AVC samples (Figure 2C). To investigate the relationship between “writers”, “erasers”, and “readers”, we analyzed pairwise correlations between the expression of these m6A regulators in AVC and discovered that positive correlations were infrequent (Figure 2D). We found a considerable association among “writers”, “erasers”, “readers”, and m6A regulators with the same role showing considerable linkage in expression. Cases of AVC with high expression of HNRNPA2B1 (a “reader” gene) exhibited downregulation of YTHDF1 (a “reader” gene), YTHDF2 (a “reader” gene), LRPPRC (a “reader” gene), FTO (an “eraser” gene), RBMX, and IGFBP1, and upregulation of WTAP (a “writer” gene) and YTHDC1 (a “reader” gene), and did not affect the expression of other genes. Cases of AVC with elevated expression of YTHDC2 (a “reader” gene) showed downregulation of YTHDF2 (a “reader” gene) and IGFBP1, and upregulation of METTL14 (a “writers” gene), YTHDC1 (a “reader” gene), HNRNPC (a “reader” gene), FTO (an “eraser” gene), and RBMX, but it did not affect the expression of other genes. The expression of the “writer” genes METTL14, RBM15, and RBM15B presented a similar trend as that of FTO (an “eraser” gene). Furthermore, variations in FMR1 and ZC3H13 expression had no impact on the expression of other genes. Hence, crosstalk between distinct m6A regulators could be critical for generating m6A modification patterns in AVC patients. We evaluated the expression of 20 m6A regulators in various m6A subtypes and disease subtypes post unsupervised clustering (Figure 2E,2F) and found that the expression of YTHDC2, IGF2BP1, RBMX, and FTO increased in the cluster B compared to their expression in the cluster A, and the expression of HNRNPA2B1 increased in the cluster A compared to its expression in the cluster B (P<0.05).
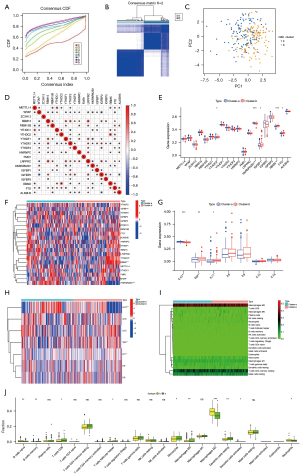
Regulation of tissue IME by m6A modification patterns in AVC
We used CIBERSORT to analyze the variations in immune cell infiltration between the 2 m6A alteration patterns. We found that cluster A was strongly related to immunological activation. Subsequent analyses of immune infiltration revealed that IL-10 expression was considerably elevated (P<0.001) in cluster A (Figure 2G,2H), and cluster A was considerably enriched in infiltrating innate immune cells, including memory B cells (P<0.01), CD8 T cells (P<0.05), T follicular helper cells (P<0.05), monocytes (P<0.05), M0 macrophages (P<0.01), and resting DCs (Figure 2I,2J). Meanwhile, naïve B cells (P<0.01), M2 macrophages (P<0.001), activated DCs (P<0.05), and neutrophils (P<0.05) were significantly abundant in cluster B. Additionally, we observed increased expression of TNF (P<0.01) and IL-11 (P<0.05) in cluster B. Significant differences in immune cell and inflammatory factor infiltration between the 2 m6A clusters suggested that m6A methylation modifications might affect the immunological milieu of the AVC tissue.
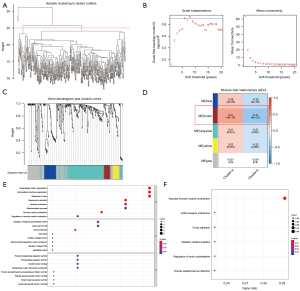
Features of key m6A modules
We utilized the “limma” package to identify the DEGs in the 2 m6A clusters. Subsequently, the first 3,000 DEGs with the lowest P value were filtered out. Following that, a WGCNA was built. After evaluating the eigengenes of each module, clustering the modules, and combining adjacent modules into new modules (Figure 3A-3C), 5 modules were acquired (Figure 3D). The brown module showed the strongest association with the m6A cluster classification phenotype (Table S3). The R package clusterProfiler was utilized to assess the genes in the brown module for function and cascade information. As illustrated in Figure 3D, the genes in the brown module were significantly enriched in BP (Figure 3D), including response to estradiol, extracellular matrix (ECM), extracellular structure organization, response to drugs, smooth muscle contraction, and muscle contraction (Figure 3E). The CCs were significantly enriched in myosin filament, collagen-comprising ECM structural constituent, and apical part of the cell (Figure 3E). The MFs were significantly enriched in protein phosphatase and phosphatase regulator activity, growth factor binding, and ECM structural component (Figure 3E). Finally, the KEGG pathways were significantly enriched in vascular smooth muscle contraction and ECM (Figure 3F).
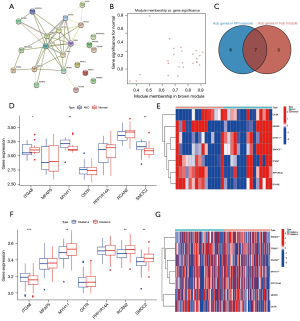
Seven key genes were linked with the AVC characteristics
To identify the key genes in important module, a PPIN was constructed, followed by analysis of 25 associated genes in the brown module (Figure 4A). After excluding non-interacting genes, the PPIN identified 13 hub genes. Genes with a gene significance above 0.3 and module membership above 0.6 were evaluated as important genes in the brown module, with 10 genes in total (Figure 4B). Next, the 10 important genes and 13 hub genes were intersected to obtain 7 key genes (Figure 4C). We found considerable differences in the expression of the 7 key genes (associated with diseases) between normal and AVC tissues using the previously merged dataset (GSE51472 and GSE12644) and found that the expression of ITGA8, MYH11, and SMOC2 was considerably elevated in AVC tissues relative to that in the normal tissues, yet the expression of the other 4 genes was indifferent (Figure 4D,4E; P<0.05). Furthermore, we found that 7 key genes exhibited significant differences in the m6A cluster categorization based on the GSE102249 dataset. The 7 key genes were expressed at a greater level in cluster A than in cluster B (Figure 4F,4G; P<0.05). Additionally, compared to cluster A, MYH11, SMOC2, and RCAN2 were considerably higher in cluster B, but ITGA8 expression was significantly higher in cluster A (Figure 4F,4G; P<0.05).
Consistent cluster analysis of GSE102249 based on 7 key genes
The AVC patients were categorized into various genomic subtypes using unsupervised clustering analysis based on the expression of 7 key genes. We found that the gene clusters could be separated (Figure 5A,5B; Table S6). The majority of individuals with the cluster B subtype were assigned to gene cluster-1 (Figure 5C). Considerable variations were observed in gene cluster classification for 7 key genes (P<0.001). The expression of RCAN2 was lower in gene cluster B. The remaining 6 key genes were expressed at significantly higher levels in gene cluster-2 than in gene cluster-1 (Figure 5D,5E; P<0.05). Additionally, we evaluated the different infiltration immune cells in the 2 subgroups (Figure 5F) and found considerable variations in CD8 T cells, memory CD4 T cells (activated), T follicular helper cells, monocytes, M1 macrophages, M2 macrophages, and eosinophils (P<0.01, 0.001, 0.001, 0.001, 0.01, 0.001, and 0.05, respectively). The M1 macrophages (P<0.01), CD8 T cells (P<0.01), memory-activated CD4 T cells (P<0.001), and T follicular helper cells (P<0.001) were highly abundant in gene cluster-1. However, monocytes, M2 macrophages, eosinophils, and neutrophils (P<0.001, 0.001, and 0.05, respectively) were substantially more abundant in gene cluster-2 (Figure 5G).
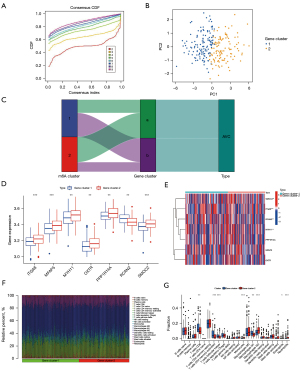
Diagnostic efficacy and pathway analysis of gene cluster subgroups
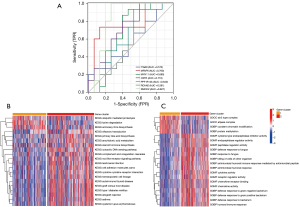
To investigate the diagnostic abilities of the 7 key genes for AVC, we performed receiver operating characteristic (ROC) curve analysis and found that SMOC2 might be the core gene with the best diagnostic effectiveness (Figure 6A). Also, the pathways in which distinct gene clusters may contribute were identified. The KEGG pathway heat map revealed that ubiquitin-mediated proteolysis, lysine degradation, and aminoacyl-tRNA biosynthesis were significantly enriched in gene cluster-1; olfactory transduction, primary bile acid biosynthesis, and arachidonic acid metabolism were significantly enriched in gene cluster-2. The GO pathway heat map showed that the sin3 type complex, ATPase complex, GOBP covalent chromatin modification, and protein methylation were significantly enriched in the significant gene cluster-1; cysteine-type endopeptidase inhibitor, endopeptidase regulator, and peptidase regulator activity were significantly enriched in the significant gene cluster-2 (Figure 6B,6C).
Discussion
As most studies on disease-related m6A modification have focused on many tumors, the impact of m6A regulators on the IME in AVC tissue may help us to understand disease development and disease-related immune infiltration and to establish more effective therapeutic regimens. Xu et al. (46) found that IL-18 stimulates M1 macrophage-mediated erythrophagocytosis and erythrocyte breakdown by activating HO-1 and FPN through p38 and Erk1/2 in a calcific microenvironment. Additional, recent findings demonstrate an adaptive immune response functions in AVC by activating circulating CD8+ cells, their clonal proliferation and differentiation to the memory-effector subtype, and exchanges of these T-cell clones between the peripheral blood and valve (47). In addition, IFN-γ from CD8+CD28 T cells may also promote calcification (47). Activated effector T cells and their cytokine production may impair osteoclast activity, increasing valvular calcium buildup. There is strong evidence that m6A regulates transcription and protein expression through splicing, translation, degradation and export, affects the activity of cancer cells and/or stromal cells and characterizes the IME (48). At present, the relationship between m6A and the IME is mainly studied in tumors, while the study on AVC is very limited. Therefore, in this study, we can further explore new therapeutic targets by analyzing the relationship between m6A-related genes and immune cells in tissues by bioinformatics.
The expression profile of common m6A regulators in calcified and normal aortic tissues was first elucidated. In the AVC samples, 2 “writers” (METTL14 and RBM15B) and 1 “reader” (HNRNPC) exhibited statistical significance, indicating their important role in AVC. The m6A methyltransferases contribute to the catalysis of the m6A modification (37). Inflammatory development is mediated by the m6A regulators. However, only 1 study found a link between m6A regulators and AVC disease development, and it was reported that METTL3 (a “writer” gene) is highly expressed in human calcified aortic valves relative to normal valves and promotes osteogenic differentiation of human aortic VICs via the m6A-YTHDF2-dependent pathway (14). A homolog of METTL3, METTL14 is a novel family of m6A “readers” (49), which is an RNA-binding scaffold that can recognize substrate RNAs (50) and enhance the proliferative and invasive potential of atherosclerotic vascular endothelial cells (51), which decrease vascular healing function (52). Histopathological analysis revealed that the initial phase of AVC is characterized by an aggressive inflammatory response similar to that seen in atherosclerotic lesions, with processes ranging from lipid deposition to infiltration of inflammatory cells including macrophages and T cells, and ultimately, destruction of the basement membrane (53). Therefore, METTL14 may affect the progression of AVC through an atherosclerosis-like pathway. Additionally, calcification of valve tissue might be the result of osteogenic differentiation of VICs and lead to osteogenic calcification and the formation of bone-like structures (54). Tian et al. showed that the m6A modification in the formation of AVC might be a key regulator involved in the osteogenic or osteoclastic processes of bone. For example, significantly elevated expression of m6A methyltransferases (METTL3 and METTL14) was found in bone mesenchymal stem cells undergoing osteogenic induction (55). Thus, METTL14 might be closely associated with the aortic valve osteocalcification phase. However, Zhou et al. (14) found no significant difference in METTL14 between normal and disease tissues in the progression of AVC. Interestingly, in our analysis, METTL14 showed low expression in AVC tissues; thus, the mechanism of action of METTL14 is not yet known, and it may be related to the regulation of various factors, which needs to be further explored. Additionally, Zhou et al. (14) found that by suppressing TWIST1 via an m6A-YTHDF2-dependent mechanism, METTL3 enhances osteogenic differentiation of human aortic VICs. In this study, we found that YTHDC1, YTHDC2, and YTHDF1 differed significantly between normal and AVC tissues; YTHDF2 and the genes mentioned above are all members of the YTH family. Thus, the YTH family might also contribute to the progression of AVC. In vitro, FTO efficiently eliminated the m6A modification of mRNA (56). Recently, some studies have found strong associations between FTO and hypertension (57), diabetes (58,59), obesity (60,61), non-alcoholic fatty liver disease (62,63), atherosclerosis (64), myocardial infarction (38,65), ischemic reperfusion injury (65), cardiac hypertrophy (65), and heart failure (65). Thus, FTO considerably contributes to the progression of cardiovascular diseases. It might be that FTO is also involved in the process of AVC. Our study revealed that the expression landscape of m6A regulators in normal and AVC patients is highly heterogeneous. The precise molecular mechanisms by which m6A regulators modulate AVC progression, however, remain unknown.
As a reversible RNA methylation process, m6A is inserted by the “writer” and deleted by the “eraser”, suggesting that they may perform antagonistic functions (66). However, our study indicated a positive correlation between the m6A writer and eraser and the frontiers of cell and developmental biology. The FTO group contains the “author” genes RBM15, RBM15B, METL14, and METL16 (a kind of “eraser” gene). Previous studies have indicated that the m6A writers that increase the amount of RNA m6A are oncogenes (67,68). The targets of “erasers” and “writers” described in various studies are distinct, suggesting that disease progression involves several processes and regulatory factors other than the “writer” and “eraser”. Furthermore, the post-transcriptional role of m6A “readers” might partly explain why the “eraser” and “writer” have contradictory roles. Future studies should determine the influence of m6A “writers” and “erasers” on the same RNA to evaluate the relative contribution of other regulatory factors in m6A modification.
In recent decades, m6A regulators have been identified in disease-associated immune cell filtration. By removing the m6A alteration, ALKBH5 reduces the antiviral natural response (69). Additionally, METTL3 might be used as an anti-inflammatory target to methylate the STAT1 mRNA and promote M1 macrophage polarization (70). The m6A reader YTHDF2 modulates the inflammatory reaction induced by lipopolysaccharides in macrophages (71). Knockout of FTO induces the reduction of PPAR-γ and STAT1 transcripts by triggering mRNA degradation (YTHDF-mediated) and attenuating M1/M2 activation (72). The immuno-infiltration analysis identified that M0 macrophages were expressed at a higher level in AVC tissues than in healthy tissues, that demonstrating that m6A regulators might affect inflammatory processes through macrophage recruitment. Furthermore, Hu et al. showed that the absence of FTO resulted in the conversion of macrophages to M1-type pro-inflammatory macrophages (73). These results suggested that FTO agonists might be potential anti-inflammatory targets. We found that FTO was expressed at low levels in AVC tissues and highly expressed in M0 macrophages, which is consistent with the results of Hu et al. Additionally, the data from AVC samples were typed by m6A expression matrix and then subjected to immuno-infiltration analysis. We found that cluster A, with elevated expression of m6A regulators, was considerably more abundant in innate immune cell infiltration, including memory B cells, CD8 T cells, T follicular helper cells, monocytes, M0 macrophages, and resting DCs, whereas, cluster B, with decreased expression of m6A regulators, was enriched in the infiltrative pattern of innate immune cells, including naïve B cells, M2 macrophages, activated DCs, and neutrophils. Thus, m6A methylation modification can considerably elevate the expression of innate immune cells linked to inflammatory processes. Cluster A had higher IL-10 expression, whereas cluster B had higher TNF and IL-1 expression. This indicated that modifying m6A methylation might affect tissue inflammatory factors in AVC. Our results on m6A regulators, however, are restricted to bioinformatics, and more research is needed to determine the exact molecular cascade through which the m6A regulator triggers immune cell infiltration in AVC. From the perspective of a biological process, GO enrichment analysis revealed enrichment in response to estradiol, ECM organization, extracellular structure organization, drug response, smooth muscle contraction, and muscle contraction. It was primarily concentrated in myosin filaments, collagen-containing ECM, and the apical region of the cell for CCs. It was primarily concentrated in phosphatase regulator activity, protein phosphatase regulator activity, growth factor binding, and ECM structural components for MFs. Further, AVC was found to be closely related to vascular smooth muscle contraction and the ECM-receptor interaction signaling pathway in the KEGG pathway analysis, which was consistent with the results of another study (74).
Next, we identified 7 disease-related hub genes utilizing key modules and discovered that the expression of ITGA8, MYH11, and SMOC2 was considerably higher in AVC tissues than in the healthy tissues. The transmembrane cell-surface receptor, ITGA8, is a member of the alpha integrin family (75). A strong association between ITGA8 and tumor development has been reported previously. For example, low ITGA8 expression is related to a poor overall survival prognosis in patients with clear cell renal cell carcinoma (76). High expression of ITGA8 causes an epithelial-mesenchymal transition in patients with early relapsed multiple myeloma that results in increased motility and invasive potential of multiple myeloma cells (77). The smooth muscle myosin, MYH11, is represented by the MYH11 gene. As a member of the myosin heavy chain family (78), MYH11 is a contractile protein that slides through actin filaments to contract muscles via adenosine triphosphate hydrolysis (79,80). Deficiency of MYH11 leads to the loss of vascular smooth muscle cells, disorganization, and hyperplasia in aortic tissues, which is one of the mechanisms causing thoracic aortic aneurysms and dissections (81,82). Thus, the association of MYH11 with AVC remains strong. The SMOC2 gene is a SPARC family member that is expressed in many tissues (83). Similarly, SMOC2 plays a role in several biological processes, including adhesion and migration of cells, angiogenesis (84), fibrosis (85), and cell proliferation (86). The involvement of SMOC2 in fibrosis during AVC needs further investigation. Our findings suggest that 7 important genes may be potential therapeutic targets for AVC. These targets may involve the development of new AVC treatments. However, further studies are needed to confirm their role in AVC progression.
The patients were categorized into various genomic subgroups based on the expression of the 7 hub genes. The CD8 T cells, memory CD4 T cells (activated), T follicular helper cells, and M1 macrophages were considerably more abundant in gene cluster-1, whereas M2 macrophages, monocytes, neutrophils, and eosinophils were considerably more abundant in gene cluster-2. The 7 key genes might play a regulatory role for immune cells in different subtypes of AVC. However, further research is needed to follow up. Furthermore, we identified the pathways in which different gene clusters may participate, which suggested that the pathways of enrichment for different subgroups of AVC were inconsistent and might be strongly related to the degree or stage of AVC lesions.
AVC has been studied for years. Ongoing clinical research examine therapies targeting recently revealed pathways. Denosumab, a human monoclonal antibody against RANKL (a member of the TNF cytokine family) that inhibits its binding to RANK (a transmembrane protein expressed on marrow stromal cells and preosteoclasts), mimics the action of osteoprotegerin, presumably limiting osteoblastic development in aortic valve tissue (87). Bisphosphonates inhibit vascular and AVC (88). They may also block AVC-related inflammatory cytokines (89). Repurposed drugs may also target AVC. Choi et al. (90) studied the effect of DPP-4 inhibitors, such as sitagliptin, on VIC calcification. NO depletion in valvular endothelial cells mimicked endothelial dysfunction, activating NF-κB pathway in VICs. AVC is a complex disease involving metabolic factors, immune cells, endothelial cells and mesenchymal cells. Inadequate in vivo model systems and in vitro method heterogeneity hinder confidence in past information. We need new methods with higher reproducibility that will increase our understanding of this disease and lead to new treatment options.
Although our findings identified potential subgroups of patients with AVC based on molecular analyses of m6A regulators and revealed the characteristics of each subtype, gene expression changes are not necessarily genetically driven and may be influenced by both genetic and environmental factors. Several additional limitations should be acknowledged. First, some clinical follow-up information was unavailable for the samples; as a result, we did not consider several important factors, such as the presence of patient comorbidities, when differentiating between subgroups. The consistency of the AVC subgroups should then be confirmed through additional analyses. The results were obtained solely through bioinformatics analysis, and they should be confirmed through additional experiments.
Conclusions
We identified a link between m6A modulators, immune infiltration, and key genes in AVC. The identification of genetic subgroups of patients with AVC has improved our understanding of the pathogenesis of AVC and has facilitated the development of potential methods for disease diagnosis, classification, and treatment.
Acknowledgments
GEO facilitated in the completion of this work. We would like to express our gratitude to the GEO network for freely sharing huge amounts of data.
Funding: This study was sponsored by funds from the Chinese National Natural Science Foundation (No. 82071673) and Qingdao West Coast New Area Science and Technology Project (No. 2020-51).
Footnote
Reporting Checklist: The authors have completed the STREGA reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-22-3627/rc
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-22-3627/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Coffey S, Roberts-Thomson R, Brown A, et al. Global epidemiology of valvular heart disease. Nat Rev Cardiol 2021;18:853-64. [Crossref] [PubMed]
- Rajamannan NM, Evans FJ, Aikawa E, et al. Calcific aortic valve disease: not simply a degenerative process: A review and agenda for research from the National Heart and Lung and Blood Institute Aortic Stenosis Working Group. Executive summary: Calcific aortic valve disease-2011 update. Circulation 2011;124:1783-91. [Crossref] [PubMed]
- Baumgartner H, Falk V, Bax JJ, et al. 2017 ESC/EACTS Guidelines for the management of valvular heart disease. Eur Heart J 2017;38:2739-91. [Crossref] [PubMed]
- Sikura KÉ, Potor L, Szerafin T, et al. Hydrogen sulfide inhibits calcification of heart valves; implications for calcific aortic valve disease. Br J Pharmacol 2020;177:793-809. [Crossref] [PubMed]
- Iung B, Vahanian A. Degenerative calcific aortic stenosis: a natural history. Heart 2012;98:iv7-13. [Crossref] [PubMed]
- Osnabrugge RL, Mylotte D, Head SJ, et al. Aortic stenosis in the elderly: disease prevalence and number of candidates for transcatheter aortic valve replacement: a meta-analysis and modeling study. J Am Coll Cardiol 2013;62:1002-12. [Crossref] [PubMed]
- Thaden JJ, Nkomo VT, Enriquez-Sarano M. The global burden of aortic stenosis. Prog Cardiovasc Dis 2014;56:565-71. [Crossref] [PubMed]
- Kostyunin AE, Yuzhalin AE, Ovcharenko EA, et al. Development of calcific aortic valve disease: Do we know enough for new clinical trials? J Mol Cell Cardiol 2019;132:189-209. [Crossref] [PubMed]
- Lindman BR, Bonow RO, Otto CM. Current management of calcific aortic stenosis. Circ Res 2013;113:223-37. [Crossref] [PubMed]
- Nishimura RA, Otto CM, Bonow RO, et al. 2017 AHA/ACC Focused Update of the 2014 AHA/ACC Guideline for the Management of Patients With Valvular Heart Disease: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation 2017;135:e1159-95. [Crossref] [PubMed]
- Salas MJ, Santana O, Escolar E, et al. Medical therapy for calcific aortic stenosis. J Cardiovasc Pharmacol Ther 2012;17:133-8. [Crossref] [PubMed]
- Cho KI, Sakuma I, Sohn IS, et al. Inflammatory and metabolic mechanisms underlying the calcific aortic valve disease. Atherosclerosis 2018;277:60-5. [Crossref] [PubMed]
- Kraler S, Blaser MC, Aikawa E, et al. Calcific aortic valve disease: from molecular and cellular mechanisms to medical therapy. Eur Heart J 2022;43:683-97. [Crossref] [PubMed]
- Zhou T, Han D, Liu J, et al. Factors influencing osteogenic differentiation of human aortic valve interstitial cells. J Thorac Cardiovasc Surg 2021;161:e163-85. [Crossref] [PubMed]
- Ducy P, Zhang R, Geoffroy V, et al. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 1997;89:747-54. [Crossref] [PubMed]
- Bäck M, Michel JB. From organic and inorganic phosphates to valvular and vascular calcifications. Cardiovasc Res 2021;117:2016-29. [Crossref] [PubMed]
- O'Brien KD, Kuusisto J, Reichenbach DD, et al. Osteopontin is expressed in human aortic valvular lesions. Circulation 1995;92:2163-8. [Crossref] [PubMed]
- Coté N, Mahmut A, Bosse Y, et al. Inflammation is associated with the remodeling of calcific aortic valve disease. Inflammation 2013;36:573-81. [Crossref] [PubMed]
- Dweck MR, Boon NA, Newby DE. Calcific aortic stenosis: a disease of the valve and the myocardium. J Am Coll Cardiol 2012;60:1854-63. [Crossref] [PubMed]
- Isoda K, Matsuki T, Kondo H, et al. Deficiency of interleukin-1 receptor antagonist induces aortic valve disease in BALB/c mice. Arterioscler Thromb Vasc Biol 2010;30:708-15. [Crossref] [PubMed]
- Meyer KD, Jaffrey SR. Rethinking m6A Readers, Writers, and Erasers. Annu Rev Cell Dev Biol 2017;33:319-42. [Crossref] [PubMed]
- Deng S, Zhang H, Zhu K, et al. M6A2Target: a comprehensive database for targets of m6A writers, erasers and readers. Brief Bioinform 2021;22:bbaa055.
- Chen XY, Zhang J, Zhu JS. The role of m6A RNA methylation in human cancer. Mol Cancer 2019;18:103. [Crossref] [PubMed]
- Zhen D, Wu Y, Zhang Y, et al. m6A Reader: Epitranscriptome Target Prediction and Functional Characterization of N 6-Methyladenosine (m6A) Readers. Front Cell Dev Biol 2020;8:741. [Crossref] [PubMed]
- Zhou Y, Yang J, Tian Z, et al. Research progress concerning m6A methylation and cancer. Oncol Lett 2021;22:775. [Crossref] [PubMed]
- Liao J, Wei Y, Liang J, et al. Insight into the structure, physiological function, and role in cancer of m6A readers-YTH domain-containing proteins. Cell Death Discov 2022;8:137. [Crossref] [PubMed]
- Winkler R, Gillis E, Lasman L, et al. m6A modification controls the innate immune response to infection by targeting type I interferons. Nat Immunol 2019;20:173-82. [Crossref] [PubMed]
- Qiu W, Zhang Q, Zhang R, et al. N6-methyladenosine RNA modification suppresses antiviral innate sensing pathways via reshaping double-stranded RNA. Nat Commun 2021;12:1582. [Crossref] [PubMed]
- Engel M, Eggert C, Kaplick PM, et al. The Role of m6A/m-RNA Methylation in Stress Response Regulation. Neuron 2018;99:389-403.e9. [Crossref] [PubMed]
- Zhou J, Wan J, Gao X, et al. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 2015;526:591-4. [Crossref] [PubMed]
- Xiang Y, Laurent B, Hsu CH, et al. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 2017;543:573-6. [Crossref] [PubMed]
- Liu N, Dai Q, Zheng G, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015;518:560-4. [Crossref] [PubMed]
- Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014;505:117-20. [Crossref] [PubMed]
- Wang X, Zhao BS, Roundtree IA, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015;161:1388-99. [Crossref] [PubMed]
- Esteve-Puig R, Bueno-Costa A, Esteller M. Writers, readers and erasers of RNA modifications in cancer. Cancer Lett 2020;474:127-37. [Crossref] [PubMed]
- Han SH, Choe J. Deciphering the molecular mechanisms of epitranscriptome regulation in cancer. BMB Rep 2021;54:89-97. [Crossref] [PubMed]
- Qin Y, Li L, Luo E, et al. Role of m6A RNA methylation in cardiovascular disease Int J Mol Med 2020;46:1958-72. (Review). [Crossref] [PubMed]
- Kumari R, Ranjan P, Suleiman ZG, et al. mRNA modifications in cardiovascular biology and disease: with a focus on m6A modification. Cardiovasc Res 2022;118:1680-92. [Crossref] [PubMed]
- Shulman Z, Stern-Ginossar N. The RNA modification N6-methyladenosine as a novel regulator of the immune system. Nat Immunol 2020;21:501-12. [Crossref] [PubMed]
- Wang H, Hu X, Huang M, et al. Mettl3-mediated mRNA m6A methylation promotes dendritic cell activation. Nat Commun 2019;10:1898. [Crossref] [PubMed]
- Han D, Liu J, Chen C, et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature 2019;566:270-4. [Crossref] [PubMed]
- Thériault S, Gaudreault N, Lamontagne M, et al. A transcriptome-wide association study identifies PALMD as a susceptibility gene for calcific aortic valve stenosis. Nat Commun 2018;9:988. [Crossref] [PubMed]
- Rysä J. Gene expression profiling of human calcific aortic valve disease. Genom Data 2016;7:107-8. [Crossref] [PubMed]
- Bossé Y, Miqdad A, Fournier D, et al. Refining molecular pathways leading to calcific aortic valve stenosis by studying gene expression profile of normal and calcified stenotic human aortic valves. Circ Cardiovasc Genet 2009;2:489-98. [Crossref] [PubMed]
- Deng X, Su R, Weng H, et al. RNA N6-methyladenosine modification in cancers: current status and perspectives. Cell Res 2018;28:507-17. [Crossref] [PubMed]
- Xu R, Zhu D, Guo J, et al. IL-18 Promotes Erythrophagocytosis and Erythrocyte Degradation by M1 Macrophages in a Calcific Microenvironment. Can J Cardiol 2021;37:1460-71. [Crossref] [PubMed]
- Winchester R, Wiesendanger M, O'Brien W, et al. Circulating activated and effector memory T cells are associated with calcification and clonal expansions in bicuspid and tricuspid valves of calcific aortic stenosis. J Immunol 2011;187:1006-14. [Crossref] [PubMed]
- Gu Y, Wu X, Zhang J, et al. The evolving landscape of N6-methyladenosine modification in the tumor microenvironment. Mol Ther 2021;29:1703-15. [Crossref] [PubMed]
- Bujnicki JM, Feder M, Radlinska M, et al. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m(6)A methyltransferase. J Mol Evol 2002;55:431-44. [Crossref] [PubMed]
- Malone T, Blumenthal RM, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J Mol Biol 1995;253:618-32. [Crossref] [PubMed]
- Zhang BY, Han L, Tang YF, et al. METTL14 regulates M6A methylation-modified primary miR-19a to promote cardiovascular endothelial cell proliferation and invasion. Eur Rev Med Pharmacol Sci 2020;24:7015-23. [PubMed]
- Chen J, Ning Y, Zhang H, et al. METTL14-dependent m6A regulates vascular calcification induced by indoxyl sulfate. Life Sci 2019;239:117034. [Crossref] [PubMed]
- Otto CM, Kuusisto J, Reichenbach DD, et al. Characterization of the early lesion of 'degenerative' valvular aortic stenosis. Histological and immunohistochemical studies. Circulation 1994;90:844-53. [Crossref] [PubMed]
- Torre M, Hwang DH, Padera RF, et al. Osseous and chondromatous metaplasia in calcific aortic valve stenosis. Cardiovasc Pathol 2016;25:18-24. [Crossref] [PubMed]
- Tian C, Huang Y, Li Q, et al. Mettl3 Regulates Osteogenic Differentiation and Alternative Splicing of Vegfa in Bone Marrow Mesenchymal Stem Cells. Int J Mol Sci 2019;20:551. [Crossref] [PubMed]
- Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011;7:885-7. [Crossref] [PubMed]
- Zotova TY, Lapaev NN, Azova MM, et al. Distribution of Polymorphisms of the Renin-Angiotensin System Genes (ACE, AGT, and AGTR1), ITGB3, and FTO in Pregnant Patients with Hypertensive Disorders. Bull Exp Biol Med 2019;167:74-8. [Crossref] [PubMed]
- Hjort R, Löfvenborg JE, Ahlqvist E, et al. Interaction Between Overweight and Genotypes of HLA, TCF7L2, and FTO in Relation to the Risk of Latent Autoimmune Diabetes in Adults and Type 2 Diabetes. J Clin Endocrinol Metab 2019;104:4815-26. [Crossref] [PubMed]
- Franzago M, Fraticelli F, Marchioni M, et al. Fat mass and obesity-associated (FTO) gene epigenetic modifications in gestational diabetes: new insights and possible pathophysiological connections. Acta Diabetol 2021;58:997-1007. [Crossref] [PubMed]
- Dina C, Meyre D, Gallina S, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet 2007;39:724-6. [Crossref] [PubMed]
- Azzam SK, Alsafar H, Sajini AA. FTO m6A Demethylase in Obesity and Cancer: Implications and Underlying Molecular Mechanisms. Int J Mol Sci 2022;23:3800. [Crossref] [PubMed]
- Hu Y, Feng Y, Zhang L, et al. GR-mediated FTO transactivation induces lipid accumulation in hepatocytes via demethylation of m6A on lipogenic mRNAs. RNA Biol 2020;17:930-42. [Crossref] [PubMed]
- Yang Z, Yu GL, Zhu X, et al. Critical roles of FTO-mediated mRNA m6A demethylation in regulating adipogenesis and lipid metabolism: Implications in lipid metabolic disorders. Genes Dis 2022;9:51-61. [Crossref] [PubMed]
- Mo C, Yang M, Han X, et al. Fat mass and obesity-associated protein attenuates lipid accumulation in macrophage foam cells and alleviates atherosclerosis in apolipoprotein E-deficient mice. J Hypertens 2017;35:810-21. [Crossref] [PubMed]
- Mathiyalagan P, Adamiak M, Mayourian J, et al. FTO-Dependent N6-Methyladenosine Regulates Cardiac Function During Remodeling and Repair. Circulation 2019;139:518-32. [Crossref] [PubMed]
- Zhou H, Yin K, Zhang Y, et al. The RNA m6A writer METTL14 in cancers: Roles, structures, and applications. Biochim Biophys Acta Rev Cancer 2021;1876:188609. [Crossref] [PubMed]
- Qian JY, Gao J, Sun X, et al. KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner. Oncogene 2019;38:6123-41. [Crossref] [PubMed]
- Lan Q, Liu PY, Haase J, et al. The Critical Role of RNA m6A Methylation in Cancer. Cancer Res 2019;79:1285-92. [Crossref] [PubMed]
- Zheng Q, Hou J, Zhou Y, et al. The RNA helicase DDX46 inhibits innate immunity by entrapping m6A-demethylated antiviral transcripts in the nucleus. Nat Immunol 2017;18:1094-103. [Crossref] [PubMed]
- Liu Y, Liu Z, Tang H, et al. The N6-methyladenosine (m6A)-forming enzyme METTL3 facilitates M1 macrophage polarization through the methylation of STAT1 mRNA. Am J Physiol Cell Physiol 2019;317:C762-75. [Crossref] [PubMed]
- Yu R, Li Q, Feng Z, et al. m6A Reader YTHDF2 Regulates LPS-Induced Inflammatory Response. Int J Mol Sci 2019;20:1323. [Crossref] [PubMed]
- Gu X, Zhang Y, Li D, et al. N6-methyladenosine demethylase FTO promotes M1 and M2 macrophage activation. Cell Signal 2020;69:109553. [Crossref] [PubMed]
- Hu F, Tong J, Deng B, et al. MiR-495 regulates macrophage M1/M2 polarization and insulin resistance in high-fat diet-fed mice via targeting FTO. Pflugers Arch 2019;471:1529-37. [Crossref] [PubMed]
- Chen J, Wang J, Jiang Y, et al. Identification of circular RNAs in human aortic valves. Gene 2018;642:135-44. [Crossref] [PubMed]
- Schnapp LM, Breuss JM, Ramos DM, et al. Sequence and tissue distribution of the human integrin alpha 8 subunit: a beta 1-associated alpha subunit expressed in smooth muscle cells. J Cell Sci 1995;108:537-44. [Crossref] [PubMed]
- Lu X, Wan F, Zhang H, et al. ITGA2B and ITGA8 are predictive of prognosis in clear cell renal cell carcinoma patients. Tumour Biol 2016;37:253-62. [Crossref] [PubMed]
- Ryu J, Koh Y, Park H, et al. Highly Expressed Integrin-α8 Induces Epithelial to Mesenchymal Transition-Like Features in Multiple Myeloma with Early Relapse. Mol Cells 2016;39:898-908. [Crossref] [PubMed]
- Matsuoka R, Yoshida MC, Furutani Y, et al. Human smooth muscle myosin heavy chain gene mapped to chromosomal region 16q12. Am J Med Genet 1993;46:61-7. [Crossref] [PubMed]
- Huxley H, Hanson J. Changes in the cross-striations of muscle during contraction and stretch and their structural interpretation. Nature 1954;173:973-6. [Crossref] [PubMed]
- Huxley AF, Niedergerke R. Structural changes in muscle during contraction; interference microscopy of living muscle fibres. Nature 1954;173:971-3. [Crossref] [PubMed]
- Zhu L, Vranckx R, Khau Van Kien P, et al. Mutations in myosin heavy chain 11 cause a syndrome associating thoracic aortic aneurysm/aortic dissection and patent ductus arteriosus. Nat Genet 2006;38:343-9. [Crossref] [PubMed]
- Pannu H, Tran-Fadulu V, Papke CL, et al. MYH11 mutations result in a distinct vascular pathology driven by insulin-like growth factor 1 and angiotensin II. Hum Mol Genet 2007;16:2453-62. [Crossref] [PubMed]
- Vannahme C, Gösling S, Paulsson M, et al. Characterization of SMOC-2, a modular extracellular calcium-binding protein. Biochem J 2003;373:805-14. [Crossref] [PubMed]
- Rocnik EF, Liu P, Sato K, et al. The novel SPARC family member SMOC-2 potentiates angiogenic growth factor activity. J Biol Chem 2006;281:22855-64. [Crossref] [PubMed]
- Luo L, Wang CC, Song XP, et al. Suppression of SMOC2 reduces bleomycin (BLM)-induced pulmonary fibrosis by inhibition of TGF-β1/SMADs pathway. Biomed Pharmacother 2018;105:841-7. [Crossref] [PubMed]
- Liu P, Lu J, Cardoso WV, et al. The SPARC-related factor SMOC-2 promotes growth factor-induced cyclin D1 expression and DNA synthesis via integrin-linked kinase. Mol Biol Cell 2008;19:248-61. [Crossref] [PubMed]
- Pawade TA, Newby DE, Dweck MR. Calcification in Aortic Stenosis: The Skeleton Key. J Am Coll Cardiol 2015;66:561-77. [Crossref] [PubMed]
- Price PA, Faus SA, Williamson MK. Bisphosphonates alendronate and ibandronate inhibit artery calcification at doses comparable to those that inhibit bone resorption. Arterioscler Thromb Vasc Biol 2001;21:817-24. [Crossref] [PubMed]
- Sansoni P, Passeri G, Fagnoni F, et al. Inhibition of antigen-presenting cell function by alendronate in vitro. J Bone Miner Res 1995;10:1719-25. [Crossref] [PubMed]
- Choi B, Lee S, Kim SM, et al. Dipeptidyl Peptidase-4 Induces Aortic Valve Calcification by Inhibiting Insulin-Like Growth Factor-1 Signaling in Valvular Interstitial Cells. Circulation 2017;135:1935-50. [Crossref] [PubMed]
(English Language Editor: J. Jones)


