Toward precision medicine in amyotrophic lateral sclerosis
Introduction
Precision medicine is an innovative approach that applies recently developed biomedical technologies to optimize and individualize treatment to the molecular drivers of an individual’s disease. It involves not creation of treatments that are unique to a patient, but rather classification of individuals into subpopulations that differ in their susceptibility to a particular disease, in the biology and/or prognosis of the disease they develop or in their response to a specific treatment (1,2). This approach of using tailored, mechanism-based therapies has been applied to cancer care and gained progressively greater impact, but it is only beginning to be considered in amyotrophic lateral sclerosis (ALS).
ALS is a neurodegenerative disease characterized by progressive deterioration mainly involving the corticospinal tract, brainstem and anterior horn cells of the spinal cord. Patients develop focal and then generalized weakness leading to paralysis. The incidence of ALS in the European population is 2–3 people per 100,000, and the overall lifetime risk of developing the condition is 1:400 (3-5). Approximately 5–10% of patients have familial ALS (FALS) and show a Mendelian pattern of inheritance; the remaining 90–95% of patients have sporadic ALS (SALS) (6). Over 60% of patients die within 3 years of presentation, usually from respiratory failure and about 10% survive for more than 10 years (7). There is no disease-modifying therapy for ALS, though riluzole slows the rate of progression and prolongs survival by 2 or 3 months (8).
ALS is a complex, multifactorial disease with variations in individual susceptibility and phenotype. The clinical and biological complexities of ALS have hindered development of effective therapeutic drugs. In this review we envision applying the key elements of precision medicine to ALS.
Comprehensive risk assessment
Genetic and environmental factors that influence susceptibility to ALS depend on multiple gene–gene and gene–environment interactions and epigenetic effects, all of which also drive phenotypic individuality. It is important to identify these modifying factors, as they could be targets for therapeutic intervention.
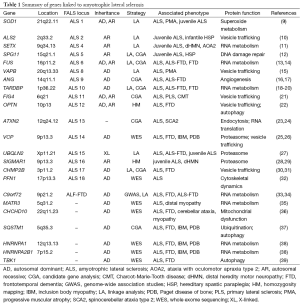
Over the last two decades, a great deal of new knowledge has been gathered on ALS, especially on its underlying genetics. To date, about 23 genes have been implicated in FALS (Table 1). Mutations in these genes account for approximately two thirds of the genetic etiology of FALS and 10% of SALS (40). Chromosome 9 open reading frame 72 (C9orf72), Cu/Zn superoxide dismutase1 (SOD1), fused in sarcoma (FUS), and TAR DNA binding protein (TARDBP) are the most common mutated genes in both FALS and SALS in various patient populations (41). Thus, there is consensus that these four genes play a causal role in ALS, whereas further evidence is required to support the roles of the other genes. Multiple genome-wide association studies (GWAS) have been performed, identifying several candidate susceptibility genes for ALS, including DPP6 (42), ELP3 (43), UNC13A (44,45), ZNF512B (46), ITPR2 and SUNC1 (47). Some of these risk genes were proposed to modify phenotype, for example, age at onset (48) and survival (49,50). However, these associations should be interpreted cautiously, as attempts to replicate the observed effects have led to either conflicting or negative results. Future GWAS should involve larger case-control cohorts and should stratify GWAS data based on different populations and well-defined clinical categories to maximize the statistical power and minimize the false discovery rate. Genome sequencing will continue to drive research in ALS genetics forward, yielding even greater insight into the genetic architecture of ALS by providing a complete catalog of rare variants. It will also allow exploration of the role of noncoding and intergenic genetic variation in the pathogenesis of this disease.
Full table
Non-genetic factors, including environmental exposure to toxins, smoking, excessive physical activity, occupation, dietary factors and changes in immunity, have been proposed as increasing risk of developing SALS. These factors may drive epigenetic changes over many years, which then induce disease onset and progression. However, the only established risk factors so far are old age and male gender. A pooled analysis of five large cohorts found that smoking is the only probable environmental risk factor for ALS, but no dose-response relationship with either pack-years or duration of smoking was found (51). Recently, the gene-time-environment model of ALS was proposed, in which the genetic component of liability, time and environmental exposures all contribute to the development of ALS (52). Environmental exposure as a risk factor for ALS, though weak, is likely to be cumulative over time, exceeding a genetic-environmental threshold in those who, at some later time, develop ALS (53). Because ALS is a rare disease, single-center studies usually lack sufficient statistical power to assess the environmental risk in ALS with known genetic backgrounds, and thus test for gene–environment interactions. Future studies of environmental risk should enroll a large sample of patients from multiple centers stratified by age of onset and known genetic risk factors.
Phenotypic classification
High phenotypic variability of ALS is observed, with regard to site of onset, age at onset, familial occurrence, type of motor neuron involvement, extent of extramotor involvement and rate of progression. Primary muscular atrophy, which involves pure lower motor neurons (LMN), and primary lateral sclerosis, which involves pure upper motor neurons (UMN), constitute the ends of a spectrum. Intermediate phenotypes, such as UMN-predominant ALS, classical ALS and LMN-predominant ALS, are considered to be different expressions of ALS. Regionally isolated variants of ALS include flail arm syndrome, which involves bilateral proximal and typically predominant LMN arm weakness, and the flail leg syndrome, characterized by often asymmetric and primarily distal LMN involvement in the lower limbs.
Advances in ALS genetics have greatly broadened the known phenotype of this disease. The discovery of TARDBP and FUS enabled recognition that ALS and frontotemporal dementia (FTD) represent overlapping clinical syndromes (13,14,18,19) and this convergence has been strengthened by the discovery of C9orf72, Ubiquilin 2 (UBQLN2) and other genes (27,33,34). It is now accepted that ALS constitutes a continuum with FTD, with pure ALS and pure FTD at the ends of this spectrum of motor neuron and frontotemporal neuron involvement. Intermediate phenotypes include ALS with behavioral impairment, ALS with cognitive impairment, and ALS-FTD (7). In addition, discovery of mutations in valosin-containing protein (VCP), Sequestosome-1 (SQSTM1), heterogeneous nuclear ribonucleoprotein A2B1 (HNRNPA2B1) and heterogeneous nuclear ribonucleoprotein A1 (HNRNPA1) in subsets of patients with ALS, FTD, inclusion body myopathy and Paget disease of the bone showed that at least some forms of ALS are part of the ‘multisystem proteinopathy’, a widespread disease process with muscle, bone and neuronal degeneration (54). Thus, there is also a spectrum of extramotor involvement in ALS, ranging from classic ALS with no or mild extramotor involvement to ALS with cognitive, extrapyramidal, cerebellar, sensory, autonomic, urinary, oculomotor, muscular or bone involvement, designated as ‘ALS with multisystem degeneration’ (7,55).
Different phenotypes of ALS may fit within a clinical and pathological continuum, or on the contrary, reflect heterogeneity of underlying pathophysiological mechanisms. Therefore, accurate disease categorization may help to explore the underlying pathophysiological mechanisms and select candidates for clinical trials.
Confirming a presymptomatic period
It is unclear whether ALS, like many neurodegenerative disorders such as Alzheimer’s, Parkinson’s and Huntington’s diseases, is characterized by a presymptomatic period. However, the fact that patients with hereditary ALS do not present clinically until mid- to late-adulthood indicates that either these mutated genes are not ‘switched on’ until later in life or that there are decades of progressive cellular compromise, eventually culminating in catastrophic decline manifesting as presentation of clinically overt ALS (53). Recent progress in the genetic basis of ALS has led to the identification of increasing numbers of asymptomatic people at genetic risk for ALS, which will help to define the presymptomatic phase of the disease.
There is a compelling body of evidence to indicate that the onset of clinical symptoms is preceded by a long presymptomatic period. Longitudinal studies reported reduced motor unit number estimation (MUNE) and increased cortical excitability 3–10 months in advance of symptom onset in SOD1 mutation carriers (56,57). It is therefore imperative to identify the sensitive biomarkers at this preclinical stage by establishing presymptomatic diagnostic tools to identify those at high risk of developing ALS. This would open a potentially important window for neuroprotective intervention that might allow rescue of dysfunctional, but not yet dead, neurons and might even enable disease prevention (53). Abnormality of upper motor neuron function was clearly shown to precede clinical deficit in ALS carrying SOD1 or C9orf72 mutations (58-60). Moreover, surprisingly homogenous miRNA alterations were found in FALS patients and asymptomatic mutation carriers (60). These findings suggested the existence of biomolecular dysfunctions at a cellular level that are insufficient to cause clinical features and that such dysfunctions are potentially present and building for years or decades prior to the onset of clinical disease (53). Further studies are needed to confirm the presymptomatic period in patients with ALS and to establish presymptomatic diagnostic tools to identify those at high risk of developing the disease.
Studying potential molecular pathways
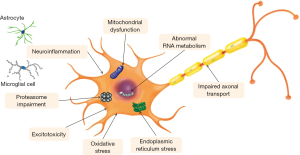
With advances in ALS genetics, several potential pathophysiological mechanisms have been implicated and proposed including oxidative stress, mitochondrial dysfunction, impairment of axonal transport, excitotoxicity, protein aggregation, endoplasmic reticulum stress, abnormal RNA processing and neuroinflammation (61) (Figure 1). Studies of ALS excitotoxicity mechanisms led to discovery of riluzole, the only FDA-proved drug for the disease. Recently, some additional molecular pathways have been identified by basic science research and therapeutic development efforts. The most notable of these include alterations in RNA metabolism and protein homeostasis.
The identification of ALS-causing mutations in the genes encoding TDP-43 and the RNA-binding protein FUS, both of which are involved in pre-mRNA splicing, RNA transport and RNA translation, led to the proposal that aberrant RNA metabolism contributes to ALS pathogenesis (13,14,18,19). Besides TDP-43 and FUS, a surprising number of proteins linked to ALS are directly or indirectly involved in RNA processing and metabolism. These proteins include TAF15, EWSR1, ANG, SETX, ELP3, ataxin-1 and -2, hnRNPA1 and hnRNPA2B1 and C9orf72 (62). TDP-43 and FUS shuttle between the nucleus and cytoplasm. In response to stressors such as starvation or oxidative stress, TDP-43 and FUS exit from the nucleus and exist primarily in the cytoplasm, where they are incorporated into stress granules and form stress granule-based aggregates. ALS-associated mutations cause a shift in localization of TDP-43 or FUS from the nucleus to the cytoplasm and increase their propensity for aggregation (63,64). This increased cytoplasmic aggregation, a gain-of-function, results in nuclear depletion of TDP-43 and FUS, inducing abnormalities in RNA processing, a loss-of-function (65-67). The identification of C9orf72 mutations as causing ALS made RNA toxicity a high profile researches focus for the disease. The C9orf72 mutations causing ALS are GGGGCC hexanucleotide repeat expansions with several hundreds or even thousands of repeats (33,34). C9orf72 mRNA levels were reduced by 50% in ALS patients with C9orf72 abnormal expansions, suggesting that the expanded allele hinders generation of mature mRNA (33,68). Thus, C9orf72 expansion may represent a loss-of-function mutation. However, the expanded hexanucleotide repeat was shown to form nuclear RNA foci in neurons in the frontal cortex and spinal cord in patients with C9orf72 mutations. Pre-mRNA containing the expansion may thus also exert a deleterious gain-of-function effect (33). Another possible mechanism of ALS pathogenesis associated with C9orf72 mutations would be repeat-associated non-ATG (RAN) translation (69). While medications to significantly reduce the gain-of-toxic function effect have not yet been discovered, targeting the production of toxic protein or RNA could achieve this aim (see discussion below).
Aggregates of mutant SOD1, TDP-43 or FUS are hallmarks of ALS. These aggregates or, more likely, their precursor oligomeric complexes, disturb normal protein homeostasis and induce cellular stress. Misfolded mutant SOD1 has toxic effects on the cell’s degradation machinery, impairing its two major components, the proteasomal pathway and autophagy, thus circumventing this protective regulatory process in the cell (70,71). Mutant SOD1 then accumulates as oligomers and later as aggregates, leading to a stress response. More evidence for interference with normal proteasomal or autophagic protein degradation as a factor in ALS pathogenesis comes from the discovery of ALS-linked mutations affecting proteins that are directly involved in proteostasis or autophagy, such as mutations in VCP, UBQLN2, SQSTM1, charged multivesicular body protein 2b (CHMP2B), optineurin (OPTN), TANK-binding kinase 1 (TBK1), TDP-43 and FUS (62). Recent studies showed that induction of autophagy might have therapeutic benefits for ALS. Molecules targeting mTOR dependent and mTOR independent autophagy pathways, including rapamycin (72,73), trehalose (74,75), spermidine, carbamazepine and tamoxifen (76), prolonged motor neuron survival or rescued motor dysfunction in mutant SOD1/TDP-43 transgenic mice, an effect correlating with increased autophagy. However, a recent phase III multicenter clinical trial reported no beneficial effects of lithium, which enhances autophagy, in ALS patient survival (77). Because most available drugs target many biological processes beyond autophagy, there is a need to explore new autophagy regulators with higher specificity and lower side effects.
Developing disease models
Laboratory models of ALS help researchers understand the basic processes of the disease, which is essential for developing new therapies. Though ALS has been modeled in cells, worms, flies, fish, mice and rats, no model is a perfect representation of the human disease, though each offers advantages for studying particular disease features. Rodents are especially important for testing potential therapies because their nervous systems are much larger and more complex than those of many other animal models. Development of transgenic animal models carrying genetic mutations identified in ALS patients has facilitated studying disease mechanisms and developing therapeutic strategies for ALS because these models recapitulate its key histopathological and biochemical features (78).
Discovery of SOD1 mutations in FALS led to generation of the first transgenic mouse model of ALS (79). SOD1 transgenic models reproduced many features of ALS, including motor deficits, reduced survival, fragmented and insoluble SOD1 aggregates, reactive gliosis and neuronal loss (79-81). SOD1 mouse models have been used extensively to study disease pathogenesis and for drug screening, but a substantial number of compounds that prolong survival and/or delay onset of paralysis in the SOD1 mouse model showed disappointing results in clinical trials. Recently, identification of TARDBP mutations led to a number of transgenic mouse models expressing either wild-type or mutant TDP-43, which have a phenotype primarily consisting of cortical abnormalities and minor lower motor neuron involvement (82,83). Transgenic mice overexpressing wild-type or mutated human FUS developed ALS-like symptoms, with hindlimb paralysis and shortened life span, along with cytoplasmic FUS aggregation (84,85). Very recently, transgenic mice with C9orf72 repeat-expansion was created, and mimicked both neuropathological and clinical C9orf72 mutated FTD/ALS phenotypes (86). These mice had neuronal loss, nuclear RNA foci, dipeptide repeat inclusions and TDP-43 pathologies in the brain, as well as such behavioral abnormalities as hyperactivity, anxiety, antisocial behavior and motor deficits. Given the high prevalence of C9orf72 mutations in FALS/ALS-FTD, transgenic C9orf72 mouse models are likely to contribute to studying the disease process and testing therapies against this form of the disease. With the number of genetic mouse models now available, compounds should be tested in models with various genetic backgrounds to determine the effects of genetic variability on therapeutic efficacy before advancement to clinical trials.
However, all of these animal models have limitations. They do not faithfully translate to human disease and each represents only a subset of FALS cases. Thus, most agents found effective in these models were not found to be of value in clinical trials (78). Reprogramming fibroblasts of ALS patients into induced pluripotent stem cells (iPSCs) appears to be a promising opportunity to develop new SALS models. These cells have been differentiated into ALS-relevant cell subtypes including motor neurons and astrocytes. Motor neurons derived from SALS patients recapitulate the major pathological features of the patients they were derived from, including TDP-43 aggregation (87). In addition, motor neurons derived from patients carrying TARDBP or C9ORF72 mutations display abnormal physiological properties (88). Thus iPSC-differentiated neurons from ALS patients offer another platform to test therapeutic candidates. The emergence of powerful gene-editing tools such as clustered regularly interspaced short palindromic repeats (CRISPR) enables introduction of specific mutations into well-characterized iPSC lines and into endogenous genes in mice. The use of endogenous mouse genes might help overcome problems arising from expression of human rather than mouse proteins (89-91). Preclinical testing in these iPSC-based models might, therefore, identify promising candidate therapies with greater effectiveness in humans (92).
Discovering biomarkers
Biomarkers would facilitate diagnosis and thus might expedite initiation of neuroprotective therapies. Biomarkers could also help select patients for enrollment in clinical trials or identify subgroups that will benefit most from certain medications. Furthermore, robust biomarkers for disease activity might also help assess drug efficacy in trials. Biomarkers that have prognostic value for survival would be of value for decision-making and planning of care. Technological developments have led to the discovery of many candidate protein-based, neurophysiological, and neuroimaging biomarkers for ALS.
Dozens of candidate protein-based biomarkers were identified in the blood and/or cerebrospinal fluid (CSF) of patients with ALS [see review in (93)]. CSF neurofilaments have become leading candidate neurochemical biomarkers of diagnostic and prognostic value (94,95). Very recently, a prospective study showed the positive predictive value of elevated levels of CSF neurofilaments for diagnosis, distinguishing between patients with ALS and neurological disorder controls (94). Neurofilaments NF-L and pNF-H were at normal levels before onset of symptoms and were increased at early symptom onset in CSF and/or serum (96). In addition, neurofilament levels correlated moderately with motor neuron disease progression and duration (94). Thus blood and CSF neurofilament levels were linked to the symptomatic phase of ALS and might, therefore, serve as objective markers of structural damage to the nervous system, a promising surrogate in disease monitoring and clinical staging of ALS and an outcome measurement for future ALS therapeutic trials (97).
A number of global physiological features can be assessed that might differentiate ALS from other diseases and enable disease progression to be monitored. MUNE enables quantification and tracing of motor unit numbers, unaffected by compensatory reinnervation during disease progression. MUNIX was shown to be more rapidly recorded and have a better reproducibility compared with other more complex MUNE methods (98). Early studies of MUNIX showed index values to be reproducible in normal subjects, whereas those with ALS tended to decline over time with disease progression. In addition, in ALS patients, the index values showed greater changes than other metrics such as compound muscle action potential amplitude, ALS functional rating scale (ALSFRS) values or forced vital capacity (99). Very recently, a multicenter prospective study performed longitudinal MUNIX measurements in multiple muscles of patients with ALS. The study demonstrated that MUNIX could track the loss of LMNs even when measuring clinically less affected muscles. In addition, MUNIX could accurately discriminate between faster or slower disease progressions. These results confirmed that MUNIX is a reliable electrophysiological biomarker to track lower motor neuron loss in ALS and might serve as a prognostic indicator (100). Electrical impedance myography, which assesses integrity and structure of a muscle, was shown to outperform other measures such as revised ALSFRS, MUNE and handheld dynamometry, in terms of its ability to detect deterioration (101), and might therefore serve as a meaningful measure of disease severity in ALS (102).
Imaging offers a noninvasive approach to biomarker discovery and disease monitoring. Voxel-based diffusion tensor imaging studies consistently showed reduced fractional anisotropy within the corticospinal tract and the corpus callosum. Voxel and surface-based MRI morphometry quantification demonstrated thinning of the primary motor cortex. However, the correlation of these imaging metrics with the absolute level of disability at the time of MRI, using ALSFRS or the estimated rate of disease progression, was inconsistent (103).
Further longitudinal studies are necessary to determine whether any protein-based candidate biomarkers, in combination with those obtained by neurophysiology and neuroimaging, will increase sensitivity and accuracy of diagnosis, help to monitor disease progression or predict prognosis in ALS (93).
Interventions tailored to individual molecular drivers
Genetic studies and outcomes from experimental models have provided the most compelling data on molecular mechanisms. Subsequent multiple clinical trials to test these proposed disease-altering interventions have been attempted but ultimately failed in treating ALS. In fact, virtually all such approaches neglected to consider the underlying clinical and biological complexities of this disease. A precision medicine approach has been overlooked until very recently, with some clinical trials aimed at specific patient populations (104). As the number of genes linked to ALS has increased, many patients have been found to harbor a mutant gene. Blocking expression of the mutant gene, therefore, stands out as a potentially definitive therapy. It might stop the complex cascade of events leading to motor neuron death before it starts. Antisense oligonucleotide (ASO) therapy has emerged as a highly promising approach for preventing mutant gene expression in neurodegenerative diseases. An interim analysis from a phase II clinical trial of ISIS-SMNRx in infants with type I spinal muscular atrophy demonstrated good tolerability and improved muscle function (105).
SOD1 targeted therapeutic strategies
Increasing evidence has indicated that the ALS-associated effects of mutant SOD1 are caused by a gain of toxic function rather than a loss of enzymatic function. Therefore, reducing concentrations of the mutant protein would be expected to slow progression of SOD1-linked ALS. In a SOD1-G93A rat model, treatment with ASOs at the time of symptom onset significantly slowed disease progression and increased survival (106). These results have led to an initial phase I clinical trial showing that intrathecal delivery of ASO targeting SOD1 (SOD1Rx) to the CNS was well tolerated by patients harboring SOD1 mutations and that CSF and plasma concentrations of ASOs were dose dependent, suggesting that intrathecal ASO delivery in patients is an effective route of delivery for ALS therapy (104). The next phase of testing will determine whether SOD1Rx can be given at a sufficiently high dosage and for a sufficiently long time to significantly reduce SOD1 protein levels, and whether such reduction will impact the course of disease in SOD1-mutated ALS. In addition to the ASO targeting SOD1, antibodies and small molecules targeting misfolded SOD1 are in preclinical testing stages (107).
C9orf72 targeted gene therapy
The SOD1Rx trial has supported pursuing potential applications of ASOs to other forms of genetically determined ALS. The likelihood that C9orf72 mutations cause a toxic gain of function makes it a promising candidate for treatment with ASO. In fact, ASOs targeting C9orf72 were shown to reduce C9orf72 pathology in iPSC-differentiated neurons from C9orf72-mutated ALS patients, including RNA aggregation, aberrant transcription factor binding, dysregulated expression of other genes, susceptibility to glutamate excitotoxicity and neuronal firing abnormalities (108-110). Furthermore, administering a mouse-specific C9orf72 ASO to the lateral ventricle of adult mice, using a single intracerebroventricular stereotactic injection, resulted in a significant reduction of C9orf72 RNA levels in the spinal cord and the brain 3 weeks after injection (110). In this study, the ASO was also detected throughout the CNS. However, no functional or behavioral alterations in strength, motor coordination, activity or anxiety were reported even at 17 weeks after the injection (110). These preliminary results indicated that the C9orf72 suppression strategy might be safe and long-lasting in animal models. Given the high prevalence of C9orf72 mutations in ALS patients of Caucasian origin, ASO could represent a promising approach for these C9orf72-mutated ALS patients.
ATXN2 targeted therapeutic strategies
Intermediate-length polyQ expansions in ATXN2 were convincingly associated with an increased risk for ALS and functional screens identified ataxin-2 as a disease-modifying factor (23,24,111). Effects of intermediate-length repeat expansions may occur not only at the protein but also at the RNA level, similar to what has been observed in other disorders caused by expanded repeat regions. Given that therapeutic strategies aimed at targeting polyQ expansions have been successfully developed and applied in other disorders, such as Huntington’s disease (112), ataxin-2 is likely to be a promising therapeutic target in ALS. ASOs and genome engineering techniques are promising approaches to target intermediate-length repeats in ATXN2. However, because manipulation of ataxin-2 may trigger unwanted side effects, such therapeutic approaches would require extensive preclinical testing (113).
TDP-43 targeted therapeutic strategies
RNA-binding proteins, such as TDP-43 and FUS, were recently shown to play a fundamental role in ALS pathogenesis. Several processes might be preferentially targeted to develop novel therapeutic or diagnostic approaches including aberrant aggregation processes, protein–protein interactions, RNA protein interactions or specific cellular pathways altered by disease (114). Compounds that decreases TDP-43 levels or reduce TDP-43 aggregation are very promising options. These include compounds activating the endoplasmic reticulum (ER) stress unfolded protein response, such as methylene blue (MB) (115), and other compounds such as cardiac glycosides, triptolide, CDK inhibitors and c-JNK inhibitors (87).
Conclusions
Together, the key elements of phenotypic classification, comprehensive risk assessment, detecting a presymptomatic period, studying potential molecular pathways, developing disease models, discovering biomarkers and tailoring interventions to molecular specifics embody a precision medicine approach. This approach should provide a strategy for optimal targeting and timing of efforts to prevent, stop or slow progression of ALS. Neuroscientists, neuropathologists and clinical researchers should work closely together to bring phenomics, genomics, proteomics, metabolomics and other unbiased approaches to bear on the problem, ensuring that future therapeutic strategies will be based on a solid molecular understanding of pathological mechanisms, identified in robust animal and cellular models.
Acknowledgements
Funding: The study was supported by the Joint Fund of Natural Science Foundation and Health Industrial Foundation (grant No. 2020174), as well as Key Clinical Specialty Discipline Construction Program of Fujian and Nation P.RC.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015;372:793-5. [PubMed]
- Montine TJ, Montine KS. Precision medicine: clarity for the clinical and biological complexity of Alzheimer's and Parkinson's diseases. J Exp Med 2015;212:601-5. [PubMed]
- Cronin S, Hardiman O, Traynor BJ. Ethnic variation in the incidence of ALS: a systematic review. Neurology. 2007;68:1002-7. [PubMed]
- Logroscino G, Traynor BJ, Hardiman O, et al. Incidence of amyotrophic lateral sclerosis in Europe. J Neurol Neurosurg Psychiatry. 2010;81:385-90. [PubMed]
- Chiò A, Mora G, Calvo A, et al. Epidemiology of ALS in Italy: a 10-year prospective population-based study. Neurology 2009;72:725-31. [PubMed]
- Chiò A, Traynor BJ, Lombardo F, et al. Prevalence of SOD1 mutations in the Italian ALS population. Neurology 2008;70:533-7. [PubMed]
- Swinnen B, Robberecht W. The phenotypic variability of amyotrophic lateral sclerosis. Nat Rev Neurol 2014;10:661-70. [PubMed]
- Miller RG, Mitchell JD, Moore DH. Riluzole for amyotrophic lateral sclerosis (ALS)/motor neuron disease (MND). Cochrane Database Syst Rev 2012;3:CD001447. [PubMed]
- Rosen DR, Siddique T, Patterson D, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993;362:59-62. [PubMed]
- Hadano S, Hand CK, Osuga H, et al. A gene encoding a putative GTPase regulator is mutated in familial amyotrophic lateral sclerosis 2. Nat Genet 2001;29:166-73. [PubMed]
- Chen YZ, Bennett CL, Huynh HM, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4). Am J Hum Genet 2004;74:1128-35. [PubMed]
- Daoud H, Zhou S, Noreau A, et al. Exome sequencing reveals SPG11 mutations causing juvenile ALS. Neurobiol Aging 2012;33:839.e5-9.
- Kwiatkowski TJ Jr, Bosco DA, Leclerc AL, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 2009;323:1205-8. [PubMed]
- Vance C, Rogelj B, Hortobagyi T, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 2009;323:1208-11. [PubMed]
- Nishimura AL, Mitne-Neto M, Silva HC, et al. A mutation in the vesicle-trafficking protein VAPB causes late-onset spinal muscular atrophy and amyotrophic lateral sclerosis. Am J Hum Genet 2004;75:822-31. [PubMed]
- Greenway MJ, Andersen PM, Russ C, et al. ANG mutations segregate with familial and 'sporadic' amyotrophic lateral sclerosis. Nat Genet 2006;38:411-3. [PubMed]
- van Es MA, Diekstra F, Veldink J, et al. A case of ALS-FTD in a large FALS pedigree with a K17I ANG mutation. Neurology 2009;72:287-8. [PubMed]
- Sreedharan J, Blair IP, Tripathi VB, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 2008;319:1668-72. [PubMed]
- Kabashi E, Valdmanis PN, Dion P, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet 2008;40:572-4. [PubMed]
- Gitcho MA, Baloh RH, Chakraverty S, et al. TDP-43 A315T mutation in familial motor neuron disease. Ann Neurol 2008;63:535-8. [PubMed]
- Chow CY, Landers JE, Bergren SK, et al. Deleterious variants of FIG4, a phosphoinositide phosphatase, in patients with ALS. Am J Hum Genet 2009;84:85-8. [PubMed]
- Maruyama H, Morino H, Ito H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature 2010;465:223-6. [PubMed]
- Elden AC, Kim HJ, Hart MP, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 2010;466:1069-75. [PubMed]
- Van Damme P, Veldink JH, van Blitterswijk M, et al. Expanded ATXN2 CAG repeat size in ALS identifies genetic overlap between ALS and SCA2. Neurology 2011;76:2066-72. [PubMed]
- Johnson JO, Mandrioli J, Benatar M, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 2010;68:857-64. [PubMed]
- Watts GD, Wymer J, Kovach MJ, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet 2004;36:377-81. [PubMed]
- Deng HX, Chen W, Hong ST, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 2011;477:211-5. [PubMed]
- Al-Saif A, Al-Mohanna F, Bohlega S. A mutation in sigma-1 receptor causes juvenile amyotrophic lateral sclerosis. Ann Neurol 2011;70:913-9. [PubMed]
- Li X, Hu Z, Liu L, et al. A SIGMAR1 splice-site mutation causes distal hereditary motor neuropathy. Neurology 2015;84:2430-7. [PubMed]
- Parkinson N, Ince PG, Smith MO, et al. ALS phenotypes with mutations in CHMP2B (charged multivesicular body protein 2B). Neurology 2006;67:1074-7. [PubMed]
- Skibinski G, Parkinson NJ, Brown JM, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet 2005;37:806-8. [PubMed]
- Wu CH, Fallini C, Ticozzi N, et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature 2012;488:499-503. [PubMed]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 2011;72:245-56. [PubMed]
- Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 2011;72:257-68. [PubMed]
- Johnson JO, Pioro EP, Boehringer A, et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci 2014;17:664-6. [PubMed]
- Bannwarth S, Ait-El-Mkadem S, Chaussenot A, et al. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain 2014;137:2329-45. [PubMed]
- Fecto F, Yan J, Vemula SP, et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol 2011;68:1440-6. [PubMed]
- Kim HJ, Kim NC, Wang YD, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 2013;495:467-73. [PubMed]
- Cirulli ET, Lasseigne BN, Petrovski S, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science 2015;347:1436-41. [PubMed]
- Renton AE, Chio A, Traynor BJ. State of play in amyotrophic lateral sclerosis genetics. Nat Neurosci 2014;17:17-23. [PubMed]
- Chiò A, Calvo A, Mazzini L, et al. Extensive genetics of ALS: a population-based study in Italy. Neurology 2012;79:1983-9. [PubMed]
- van Es MA, van Vught PW, Blauw HM, et al. Genetic variation in DPP6 is associated with susceptibility to amyotrophic lateral sclerosis. Nat Genet 2008;40:29-31. [PubMed]
- Simpson CL, Lemmens R, Miskiewicz K, et al. Variants of the elongator protein 3 (ELP3) gene are associated with motor neuron degeneration. Hum Mol Genet 2009;18:472-81. [PubMed]
- van Es MA, Veldink JH, Saris CG, et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat Genet 2009;41:1083-7. [PubMed]
- Shatunov A, Mok K, Newhouse S, et al. Chromosome 9p21 in sporadic amyotrophic lateral sclerosis in the UK and seven other countries: a genome-wide association study. Lancet Neurol 2010;9:986-94. [PubMed]
- Iida A, Takahashi A, Kubo M, et al. A functional variant in ZNF512B is associated with susceptibility to amyotrophic lateral sclerosis in Japanese. Hum Mol Genet 2011;20:3684-92. [PubMed]
- Chiò A, Schymick JC, Restagno G, et al. A two-stage genome-wide association study of sporadic amyotrophic lateral sclerosis. Hum Mol Genet 2009;18:1524-32. [PubMed]
- Ahmeti KB, Ajroud-Driss S, Al-Chalabi A, et al. Age of onset of amyotrophic lateral sclerosis is modulated by a locus on 1p34.1. Neurobiol Aging 2013;34:357.e7-19.
- Chiò A, Mora G, Restagno G, et al. UNC13A influences survival in Italian amyotrophic lateral sclerosis patients: a population-based study. Neurobiol Aging 2013;34:357.e1-5.
- Tetsuka S, Morita M, Iida A, et al. ZNF512B gene is a prognostic factor in patients with amyotrophic lateral sclerosis. J Neurol Sci 2013;324:163-6. [PubMed]
- Wang H, O'Reilly EJ, Weisskopf MG, et al. Smoking and risk of amyotrophic lateral sclerosis: a pooled analysis of 5 prospective cohorts. Arch Neurol 2011;68:207-13. [PubMed]
- Al-Chalabi A, Hardiman O. The epidemiology of ALS: a conspiracy of genes, environment and time. Nat Rev Neurol 2013;9:617-28. [PubMed]
- Eisen A, Kiernan M, Mitsumoto H, et al. Amyotrophic lateral sclerosis: a long preclinical period? J Neurol Neurosurg Psychiatry 2014;85:1232-8. [PubMed]
- Taylor JP. Multisystem proteinopathy: Intersecting genetics in muscle, bone, and brain degeneration. Neurology 2015;85:658-60. [PubMed]
- Benatar M, Wuu J, Fernandez C, et al. Motor neuron involvement in multisystem proteinopathy: implications for ALS. Neurology 2013;80:1874-80. [PubMed]
- Aggarwal A, Nicholson G. Detection of preclinical motor neurone loss in SOD1 mutation carriers using motor unit number estimation. J Neurol Neurosurg Psychiatry 2002;73:199-201. [PubMed]
- Vucic S, Nicholson GA, Kiernan MC. Cortical hyperexcitability may precede the onset of familial amyotrophic lateral sclerosis. Brain 2008;131:1540-50. [PubMed]
- Ng MC, Ho JT, Ho SL, et al. Abnormal diffusion tensor in nonsymptomatic familial amyotrophic lateral sclerosis with a causative superoxide dismutase 1 mutation. J Magn Reson Imaging 2008;27:8-13. [PubMed]
- Carew JD, Nair G, Andersen PM, et al. Presymptomatic spinal cord neurometabolic findings in SOD1-positive people at risk for familial ALS. Neurology 2011;77:1370-5. [PubMed]
- Walhout R, Schmidt R, Westeneng HJ, et al. Brain morphologic changes in asymptomatic C9orf72 repeat expansion carriers. Neurology 2015;85:1780-8. [PubMed]
- Ferraiuolo L, Kirby J, Grierson AJ, Sendtner M, Shaw PJ. Molecular pathways of motor neuron injury in amyotrophic lateral sclerosis. Nat Rev Neurol 2011;7:616-30. [PubMed]
- Robberecht W, Philips T. The changing scene of amyotrophic lateral sclerosis. Nat Rev Neurosci 2013;14:248-64. [PubMed]
- Bosco DA, Lemay N, Ko HK, et al. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet 2010;19:4160-75. [PubMed]
- Ito D, Suzuki N. Conjoint pathologic cascades mediated by ALS/FTLD-U linked RNA-binding proteins TDP-43 and FUS. Neurology 2011;77:1636-43. [PubMed]
- Tollervey JR, Curk T, Rogelj B, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci 2011;14:452-8. [PubMed]
- Polymenidou M, Lagier-Tourenne C, Hutt KR, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat Neurosci 2011;14:459-68. [PubMed]
- Sun S, Ling SC, Qiu J, et al. ALS-causative mutations in FUS/TLS confer gain and loss of function by altered association with SMN and U1-snRNP. Nat Commun 2015;6:6171. [PubMed]
- Gijselinck I, Van Langenhove T, van der Zee J, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol 2012;11:54-65. [PubMed]
- Pearson CE. Repeat associated non-ATG translation initiation: one DNA, two transcripts, seven reading frames, potentially nine toxic entities! PLoS Genet 2011;7:e1002018. [PubMed]
- Bendotti C, Marino M, Cheroni C, et al. Dysfunction of constitutive and inducible ubiquitin-proteasome system in amyotrophic lateral sclerosis: implication for protein aggregation and immune response. Prog Neurobiol 2012;97:101-26. [PubMed]
- Chen S, Zhang X, Song L, Le W. Autophagy dysregulation in amyotrophic lateral sclerosis. Brain Pathol 2012;22:110-6. [PubMed]
- Caccamo A, Majumder S, Deng JJ, Bai Y, Thornton FB, Oddo S. Rapamycin rescues TDP-43 mislocalization and the associated low molecular mass neurofilament instability. J Biol Chem 2009;284:27416-24. [PubMed]
- Ryu HH, Jun MH, Min KJ, et al. Autophagy regulates amyotrophic lateral sclerosis-linked fused in sarcoma-positive stress granules in neurons. Neurobiol Aging 2014;35:2822-31. [PubMed]
- Castillo K, Nassif M, Valenzuela V, et al. Trehalose delays the progression of amyotrophic lateral sclerosis by enhancing autophagy in motoneurons. Autophagy 2013;9:1308-20. [PubMed]
- Zhang X, Chen S, Song L, et al. MTOR-independent, autophagic enhancer trehalose prolongs motor neuron survival and ameliorates the autophagic flux defect in a mouse model of amyotrophic lateral sclerosis. Autophagy 2014;10:588-602. [PubMed]
- Wang IF, Guo BS, Liu YC, et al. Autophagy activators rescue and alleviate pathogenesis of a mouse model with proteinopathies of the TAR DNA-binding protein 43. Proc Natl Acad Sci U S A 2012;109:15024-9. [PubMed]
- Morrison KE, Dhariwal S, Hornabrook R, et al. Lithium in patients with amyotrophic lateral sclerosis (LiCALS): a phase 3 multicentre, randomised, double-blind, placebo-controlled trial. Lancet Neurol 2013;12:339-45. [PubMed]
- Philips T, Rothstein JD. Rodent Models of Amyotrophic Lateral Sclerosis. Curr Protoc Pharmacol 2015;69:5.67.1-21.
- Gurney ME, Pu H, Chiu AY, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 1994;264:1772-5. [PubMed]
- Bruijn LI, Becher MW, Lee MK, et al. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron 1997;18:327-38. [PubMed]
- Wong PC, Pardo CA, Borchelt DR, et al. An adverse property of a familial ALS-linked SOD1 mutation causes motor neuron disease characterized by vacuolar degeneration of mitochondria. Neuron 1995;14:1105-16. [PubMed]
- Arnold ES, Ling SC, Huelga SC, et al. ALS-linked TDP-43 mutations produce aberrant RNA splicing and adult-onset motor neuron disease without aggregation or loss of nuclear TDP-43. Proc Natl Acad Sci U S A 2013;110:E736-45. [PubMed]
- Wils H, Kleinberger G, Janssens J, et al. TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci U S A 2010;107:3858-63. [PubMed]
- Mitchell JC, McGoldrick P, Vance C, et al. Overexpression of human wild-type FUS causes progressive motor neuron degeneration in an age- and dose-dependent fashion. Acta Neuropathol 2013;125:273-88. [PubMed]
- Qiu H, Lee S, Shang Y, et al. ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J Clin Invest 2014;124:981-99. [PubMed]
- Chew J, Gendron TF, Prudencio M, et al. Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science 2015;348:1151-4. [PubMed]
- Burkhardt MF, Martinez FJ, Wright S, et al. A cellular model for sporadic ALS using patient-derived induced pluripotent stem cells. Mol Cell Neurosci 2013;56:355-64. [PubMed]
- Lenzi J, De Santis R, de Turris V, et al. ALS mutant FUS proteins are recruited into stress granules in induced pluripotent stem cell-derived motoneurons. Dis Model Mech 2015;8:755-66. [PubMed]
- Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339:819-23. [PubMed]
- Yang H, Wang H, Shivalila CS, et al. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 2013;154:1370-9. [PubMed]
- Wang H, Yang H, Shivalila CS, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013;153:910-8. [PubMed]
- Ittner LM, Halliday GM, Kril JJ, et al. FTD and ALS--translating mouse studies into clinical trials. Nat Rev Neurol 2015;11:360-6. [PubMed]
- Bowser R, Turner MR, Shefner J. Biomarkers in amyotrophic lateral sclerosis: opportunities and limitations. Nat Rev Neurol 2011;7:631-8. [PubMed]
- Steinacker P, Feneberg E, Weishaupt J, et al. Neurofilaments in the diagnosis of motoneuron diseases: a prospective study on 455 patients. J Neurol Neurosurg Psychiatry 2016;87:12-20. [PubMed]
- Lu CH, Macdonald-Wallis C, Gray E, et al. Neurofilament light chain: A prognostic biomarker in amyotrophic lateral sclerosis. Neurology. 2015;84:2247-57. [PubMed]
- Weydt P, Oeckl P, Huss A, et al. Neurofilaments levels as biomarkers in asymptomatic and symptomatic familial ALS. Ann Neurol 2015. [Epub ahead of print]. [PubMed]
- Turner MR, Gray E. Are neurofilaments heading for the ALS clinic? J Neurol Neurosurg Psychiatry 2016;87:3-4. [PubMed]
- Furtula J, Johnsen B, Christensen PB, et al. MUNIX and incremental stimulation MUNE in ALS patients and control subjects. Clin Neurophysiol 2013;124:610-8. [PubMed]
- Neuwirth C, Nandedkar S, Stalberg E, et al. Motor Unit Number Index (MUNIX): reference values of five different muscles in healthy subjects from a multi-centre study. Clin Neurophysiol 2011;122:1895-8. [PubMed]
- Neuwirth C, Barkhaus PE, Burkhardt C, et al. Tracking motor neuron loss in a set of six muscles in amyotrophic lateral sclerosis using the Motor Unit Number Index (MUNIX): a 15-month longitudinal multicentre trial. J Neurol Neurosurg Psychiatry 2015;86:1172-9. [PubMed]
- Rutkove SB, Caress JB, Cartwright MS, et al. Electrical impedance myography as a biomarker to assess ALS progression. Amyotroph Lateral Scler. 2012;13:439-45. [PubMed]
- Rutkove SB, Caress JB, Cartwright MS, et al. Electrical impedance myography correlates with standard measures of ALS severity. Muscle Nerve 2014;49:441-3. [PubMed]
- Bede P, Hardiman O. Lessons of ALS imaging: Pitfalls and future directions - a critical review. Neuroimage Clin 2014;4:436-43. [PubMed]
- Miller TM, Pestronk A, David W, et al. An antisense oligonucleotide against SOD1 delivered intrathecally for patients with SOD1 familial amyotrophic lateral sclerosis: a phase 1, randomised, first-in-man study. Lancet Neurol 2013;12:435-42. [PubMed]
- Castro D, Iannaccone ST. Spinal muscular atrophy: therapeutic strategies. Curr Treat Options Neurol 2014;16:316. [PubMed]
- Smith RA, Miller TM, Yamanaka K, et al. Antisense oligonucleotide therapy for neurodegenerative disease. J Clin Invest 2006;116:2290-6. [PubMed]
- Liu HN, Tjostheim S, Dasilva K, et al. Targeting of monomer/misfolded SOD1 as a therapeutic strategy for amyotrophic lateral sclerosis. J Neurosci. 2012;32:8791-9. [PubMed]
- Donnelly CJ, Zhang PW, Pham JT, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron 2013;80:415-28. [PubMed]
- Sareen D, O'Rourke JG, Meera P, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med 2013;5:208ra149.
- Lagier-Tourenne C, Baughn M, Rigo F, et al. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci U S A 2013;110:E4530-9. [PubMed]
- Lee T, Li YR, Ingre C, et al. Ataxin-2 intermediate-length polyglutamine expansions in European ALS patients. Hum Mol Genet 2011;20:1697-700. [PubMed]
- Kay C, Skotte NH, Southwell AL, et al. Personalized gene silencing therapeutics for Huntington disease. Clin Genet 2014;86:29-36. [PubMed]
- van den Heuvel DM, Harschnitz O, van den Berg LH, et al. Taking a risk: a therapeutic focus on ataxin-2 in amyotrophic lateral sclerosis? Trends Mol Med 2014;20:25-35. [PubMed]
- Romano M, Buratti E. Targeting RNA binding proteins involved in neurodegeneration. J Biomol Screen 2013;18:967-83. [PubMed]
- Vaccaro A, Patten SA, Aggad D, et al. Pharmacological reduction of ER stress protects against TDP-43 neuronal toxicity in vivo. Neurobiol Dis 2013;55:64-75. [PubMed]