SOS response and its regulation on the fluoroquinolone resistance
Introduction
Free-living bacteria commonly face changing environments and must cope with varying condition, thus they have developed mechanisms that favour genome modifications either by transiently increasing their mutation rates, inducing re-arrangements, or by horizontal gene transfer (HGT) (1). One of the better known responses of this kind is the trigger of the SOS, by which bacteria can counteract DNA damage and promote survival to antibiotics like fluoroquinolones. The development of fluoroquinolone resistance by bacteria constitutes a remarkable bacterial success story, in which the SOS response plays an indispensable role.
SOS induction
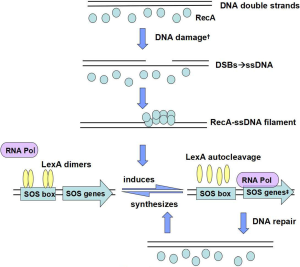
An increase in expression of the SOS genes begins when DNA is damaged, or when replication of DNA is blocked and single stranded DNA (ssDNA) accumulates. As the sole inducer of the SOS response, ssDNA mostly originates from double-strand breaks (DSBs) (2). Al Mamun argued that the SOS response was activated by DSBs and promoted mutation via transcriptional up-regulation of DNA polymerases (Pols) IV and V (3), which respectively appeared in the early and final stages of SOS induction, and Pol V was seen as the most error-prone enzyme (4). Several internal and additional external processes could trigger the SOS response, which was first identified in UV-irradiated E. coli cells and was soon also linked to other DNA-damaging factors, like mitomycin C, antibiotics, classic DNA-damaging agents, endogenous alkylating agents like nitrosated amines or S-adenosylmethionine, chromate shock, acoustic cavitation or pH levels (5), all of which would create DNA DSBs that subsequently lead to SOS induction (Figure 1). It is to be observed that Kohanski et al. have demonstrated that sublethal concentrations of some bactericidal antibiotics induce mutagenesis and that this induction correlates with an increase in reactive oxygen species (ROS), which in turn produces induction of the SOS response in 2003 (6). Nevertheless, more recently, Liu demonstrated that antibiotic exposures did not produce ROS and that lethality more likely resulted from the direct inhibition of cell-wall assembly, protein synthesis, and DNA replication on Science (7), meanwhile, another study also reported that ROS do not play a role in killing of bacterial pathogens by antibiotics (8), both of which drew diametrically opposite conclusions from former study.
SOS genes
Many bacteria are able to mount the SOS response, which involves more than 30 genes, allowing bacteria to increase DNA damage tolerance and DNA repair. Members of the SOS regulon include umuDC, recA, uvrA and dinB and many others (9). According to the extensive research on SOS response in Escherichia coli, more than 40 genes are directly regulated by LexA (10). Analyses in Bacillus subtilis have enlarged the list of LexA-regulated genes to 33 while the initial SOS network of the B. subtilis reported only five SOS-inducible genes (11), but only seven genes were found to be common between these two bacteria (12). And 16 LexA target genes have been identified in Staphylococcus aureus (13).
Moreover, it should come as no surprise that lexA itself is an SOS gene (14). the recombination and repair genes recA, recN, and ruvAB, the nucleotide excision repair genes uvrAB and uvrD, the error-prone DNA polymerase (Pol) genes dinB (encoding Pol IV) and umuDC (encoding Pol V), and DNA polymerase II, are regulated by the LexA regulon (15). Notably, the low-fidelity, error-prone repair DNA polymerases permit DNA replication across persistent DNA lesions that block the primary replicative DNA polymerase Pol III, but also promote an elevated mutation rate that generates genetic diversity and adaptation, including the evolution of antibiotic resistance. The SOS genes, however, are not all induced at the same time and to the same level, which varies due to differences in LexA binding affinity, number and location of the SOS boxes relative to the promoter, as well as promoter strength. The first genes to be induced are uvrA, uvrB, and uvrD (14).
LexA and RecA protein
Under normal conditions, LexA represses the transcription of a regulon encompassing more than 50 genes that encoding various DNA repair proteins by binding to SOS box (the region of a promoter that is recognized by LexA) in its operator, in the form of dimers (16). Zhang et al. also demonstrated that the LexA protein contained two domains: an N-terminal winged helix-turn-helix (wHTH) DNA binding domain, and a C-terminal dimerization and latent protease domain, and the relative position of the N- and C-terminal domains is highly variable (16). In the three-dimensional model of the RecA-ssDNA-ATP complexes Kovačič et al. generated, besides the catalytic C-terminal domain of LexA, its N-terminal DNA-binding domain also interacted with RecA–ssDNA filament (RecA*) (17).
Once the cell senses the presence of an increased level of DNA damage, accumulating RecA-ssDNA-ATP complexes activate LexA for autocleavage and the SOS genes are de-repressed. While the LexA protein is the repressor, The RecA protein is the inducer, working together alternate between on and off states (18). Contact with single-stranded DNA activates the coprotease activity of the RecA protein, which promotes self-cleavage of LexA, and leads to increased transcription of the SOS response regulon (19). While the repair work complete, SOS induction would be reversed due to the disappearance of the RecA filament, and the newly synthesized LexA dimers bind to SOS box. Additionally, RecA not only plays a major role in UmuD, promotion homologous recombination and the rescue of stalled replication, but is also important for control of swarming motility and the behaviour of bacteria in biofilms (20).
However, RecA binding to ssDNA is regulated. Previous study suggested that RecA in E. coli is loaded by the RecBCD and RecFOR pathways in vitro and in vivo (21). Based upon early and recent studies (22,23), RecBCD operates late in the recombination process—after initiation, strand invasion, and crossover resolution have occurred, processing double-strand ends and loading RecA onto single-stranded DNA. A previous study of RecFOR proteins and RecA protein had emphasized that the RecFOR proteins specifically target RecA protein to gapped DNA (gDNA) even in the presence of a thousand-fold excess of single-stranded DNA (24).
LexA/RecA-independent pathways
It was evident that mutations could not be completely obliterated with recA deletion in Renu Singh’s recA-deleted E. coli models (15), which has demonstrated that besides LexA/RecA dependent SOS regulatory system, there are LexA/RecA-independent pathways to trigger the SOS response. For instance, several β-lactams can induce translesion synthesis and mutagenesis by activating dinB via LexA/RecA-independent way, another example, many of the DNA repair genes of Mycobacterium tuberculosis have been shown to be DNA damage-inducible in a LexA/RecA-independent manner (25). What’s more, fluoroquinolones have also been suggested to stimulate intra- and interchromosomal recombination in E. coli through a mechanism that does not require LexA cleavage (26).
Fluoroquinolone resistance
Fluoroquinolones are commonly prescribed antimicrobial agents all over the world and have seen increasing clinical use because of their potent and broad antimicrobial activity. Unfortunately, over the 20 years that have elapsed since the introduction of fluoroquinolones, the prevalence of fluoroquinolone resistance amongst clinical isolates has become an increasingly challenge at an alarming speed. Based on a meta-analysis of Shigella in the area of Asia-Africa, resistance rate to ciprofloxacin was 0.6% during the years 1998-2000 and dramatically rose to 29.1% in 2007-2009, this 12-year period witnessed a 48.5-fold increase in resistance to ciprofloxacin (27). Acinetobacter baumannii is one of the main infectious nosocomial pathogens which lead multidrug-resistant strains worldwide, showing resistance to clinical commonly used antibiotics such as cephalosporins, carbapenems, fluoroquinolones and aminoglycosides. Another report argued that overuse of fluoroquinolone antibiotics (FQs) in medicine had promoted bacterial resistance to FQs in recent years, which had caused a huge challenge in the anti-infective therapy of Pseudomonas aeruginosa (28).
Quinolone-resistance determining region (QRDR)
Resistance to fluoroquinolones typically arises as a result of alterations in the two essential target enzymes: DNA gyrase and topoisomerase IV. DNA gyrase is the more susceptible target in which mutations are selected first in gram-negative bacteria, whereas the topoisomerase IV in gram-positive bacteria (29). Both are large, complex enzymes composed of 2 pairs of subunits. The subunits of DNA gyrase are GyrA and GyrB, encoded by the gyrA and gyrB genes, respectively. While the corresponding subunits of topoisomerase IV are ParC and ParE, encoded by the parC and parE genes, together termed the quinolone-resistance determining region (QRDR). In Enterobacteriaceae, fluoroquinolone resistance is mainly caused by point mutations in the quinolone resistance-determining region of gyrase (gyrA and gyrB) and topoisomerase (parC and parE) genes (30). Likewise, Bonomo et al. have shown that resistance to fluoroquinolone in Acinetobacter baumannii is mainly caused by mutations in the QRDRs of gyrase and topoisomerase genes (31).
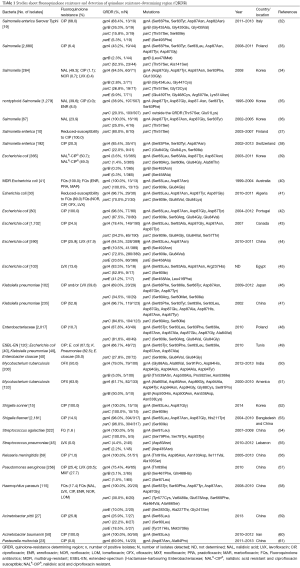
Some epidemiological surveys have been reported using polymerase chain reaction (PCR) methodologies to examine QRDRs and mutations occurred leading to the clinical fluoroquinolone resistance (Table 1), we have randomly reviewed 30 articles in total of 13,068 strains including Salmonella, Escherichia coli, Klebsiella pneumoniae, Enterobacteriaceae, Mycobacterium tuberculosis, Shigella, Streptococcus, Neisseria meningitidis, Pseudomonas aeruginosa, Haemophilus parasuis, Acinetobacter and Pasteurella multocida studied in different areas and different years. There are 4,247 (32.5%) Salmonella with the fluoroquinolones ciprofloxacin resistance changed from 0.0% to 68.0%, 2,911 (22.3%) Escherichia coli and among ciprofloxacin resistance ones, the average drug resistance rate was 30.3%, 337 Klebsiella pneumoniae strains in two reports with fluoroquinolone resistance up to more than 50.0%. Of the two studies on Enterobacteriaceae including three or more bacteria, the prevalence of ciprofloxacin resistance was 10.7% and 61.7%, respectively. The drug resistance of Mycobacterium tuberculosis [333] was observed for ofloxacin with an average prevalence of 57.0%. Given the circumstances of high resistance rate, the frequently seen point mutations in the isolates detected were gyrA Ser83Phe/Tyr/Ile and Asp87Asn/Tyr, while among parC genes, the commonly abundant mutations were Ser80Ile and Glu84Gly/Val. However, the frequencies of gyrB and parE genes were much lower in the researches we reviewed, for gyrB, mutations of Gly, Leu, Ser, Asp, Asn and Gln amino acid substitution at different codons were reported, and it may be a few more common to see Ser substituted by others in parE genes. Also, some new target gene mutations have been detected in recent years, which exhibit polymorphism of fluoroquinolone resistance.
Full table
Plasmid-mediated quinolone resistance (PMQR)
Another mechanism of fluoroquinolone resistance PMQR have also been characterized, which was first reported in a clinical isolate of Klebsiellae pneumoniae from the USA in 1988, named the qnrA (62), since then, other PMQR determinants have been detected: qnrB, qnrS, aac(6’)-Ib-cr and qepA, conferring low-level resistance to fluoroquinolone (63). It has been reported that aac(6’)-Ib-cr significantly increased the frequency of selection of chromosomal mutants upon exposure to ciprofloxacin, as for the MIC of ciprofloxacin increased by 3- to 4-fold when aac(6’)-Ib-cr was introduced into E. coli (64).
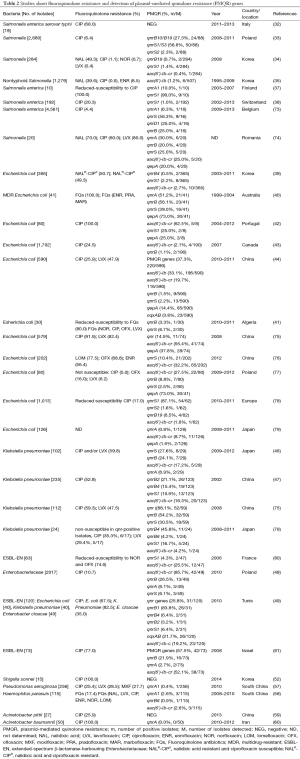
In the present study, fluoroquinolone resistance could be transferred by conjugation from all four PMQR-positive donors, suggesting that the dissemination of the PMQR determinants is mostly due to the transmission of plasmids by horizontal exchange (65). Additionally, also noteworthy is the finding that the strong association between broad-spectrum β-lactamases and qnr genes, most qnr-bearing plasmids for which sequencing is available carry a β-lactamase gene, which creates a situation ripe for the dissemination of multidrug-resistant Enterobacteriaceae (66). PMQR has so far managed to achieve global distribution in a variety of plasmid environments and bacterial genera (67), the qnrS, aac(6’)-Ib-cr and qepA genes were predominantly distributed in China (30,67-69), while the qnrB account for a moiety of PMQR genes besides others in some other countries (70-72). Also, it is believed that the presence of PMQR may facilitate the selection of QRDR mutations, resulting in higher levels of fluoroquinolone resistance. The data we concluded from several studies has shown the high prevalence of PMQR genes and the relatively resistance to fluoroquinolones in certain organisms (Table 2), the qnrB, qnrS and aac(6’)-Ib genes were at higher prevalences among the PMQR genes detected, with the positive rate of 10.4% in 2,269 isolates, 11.9% in 2,066 isolates and 16.5% in 3088 isolates, respectively. Moreover, six variant qnrB genes (qnrB1/2/4/6/10/19) and three qnrS genes (qnrS1/2/3) were identified in several studies. Besides, qnrA accounted for 5.6% in a total of 735 strains in 10 reports, efflux pump gene qepA reached 20.1% in 900 strains of 7 reports. Nevertheless, another efflux pump gene, oqxAB, only detected in two studies with 49 out of 710 was positive. No qnrC gene was reported and qnrD was detected in none but a report on Salmonella enterica. The data listed might set off alarm bells, which calls for intensify fluoroquinolone surveillance and a more cautious approach to fluoroquinolone use.
Full table
Other mechanisms
Except for the QRDR and PMQR, the other classically described mechanism of fluoroquinolone resistance operates by decreasing intracellular drug accumulation via upregulation of native efflux pumps, which has been reported in a number of Gram-negative pathogenic bacteria such as E. coli, P. Aeruginosa and Shigella dysenteriae (82). Additionally, the alterations in the composition of bacterial outer membrane proteins (OMPs) may render a strain more or less permissive towards fluoroquinolones (83).
SOS regulation
It should be noted that these reports suggest that fluoroquinolone resistance in different regions seemed to have had different characteristics; furthermore, the mutation may vary in different strains. It has been demonstrated that ciprofloxacin stimulate mutagenesis in E. coli through the induction of mutagenic DNA polymerases of the SOS system, with a 106-fold increase in mutational frequency (16). Besides, Cirz and Romesberg have recently shown that the evolution of resistance to ciprofloxacin in vivo and in vitro requires the induction of a mutation that is mediated by the cleavage of the SOS repressor LexA and the associated derepression of three specialized DNA polymerases (Pol II, Pol IV, and Pol V) (84). Afterwards, a study showed that a majority of persisters to ciprofloxacin were formed upon exposure to the antibiotic, in a manner dependent on the SOS gene network, contrary to the prevailing view of persister formation (85). Likewise, in Staphylococcus aureus, persistence and the evolution of resistance may be related to several complex regulatory networks, such as the SOS response, which modifies transcription in response to environmental stress. It has been confirmed that ciprofloxacin leads to higher recA transcription and translation as well as activation of the SOS response, which was indicated by the up-regulation of the error-prone polymerase umuC, accounting for the higher mutation frequency (86).
A research has demonstrated that fluoroquinolone-resistance phenotype of a gyrA mutation was influenced by mutations in the recA gene, by the fact that recA142 mutation caused a remarkable decrease in fluoroquinolone resistance (87). Moreover, recBCD mutations affecting recombination also reduce the level of fluoroquinolone resistance, indicating that an SOS-dependent process was acting in the repair of DNA damage (Figure 2).
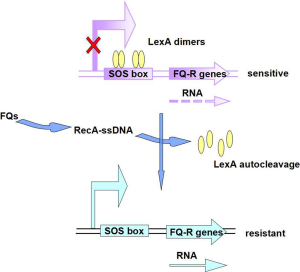
A study targeting the impact of recA on levofloxacin in Staphylococcus aureus and Escherichia coli showed that recA deletion itself resulted in a 4-fold reduction in the levofloxacin MIC, and E. coli resistance emergence was delayed by 24 h in the recA-deleted mutant, which provided useful insights into a potential target to combat the looming danger of antibiotic resistance (16). What’s more, Yim et al. stated that even at the subinhibitory concentrations employed, the older as well as newer FQs, upregulating genes involved in the SOS response, umuD, lexA, sbmC and dinP (88).
Sandra Da Re and his partners have interestingly identified the CTGTATAAAAAAACAG sequence between the +1 start site and the initiation codon of qnrB2, which is homologous to the gammaproteobacteria LexA-protein-binding site consensus, CTGTN8ACAG, suggesting that qnrB2 expression might be regulated through the SOS response in a LexA/RecA-dependent manner, and that it can be induced by ciprofloxacin, a known inducer of the SOS system (89). And via a recombinant plasmid, pPqnrB2-lacZ, they confirmed that LexA is involved in qnrB2 negative regulation, by binding to the identified motif. Since phylogenetic analysis have showed that qnrB and qnrD are closer to one another than to the other qnr determinants, qnrA, qnrS and qnrC, a potential LexA-binding site was identified upstream from the qnrD gene (89). Wang et al. have also examined that in the sequence upstream from qnrB (but not qnrA or qnrS) was a LexA binding site, and qnrB was shown to be under SOS control by demonstrating that fluoroquinolone susceptibility decreased with increasing temperature (90).
Recently, a new pentapeptide repeat proteins (PRP) protein, named SmaQnr, which shares 80% amino acid identity with QnrB1, has been reported as reducing susceptibility to fluoroquinolone when expressed in both E. coli ATCC 25922 and E. coli DH10B. Sequences upstream of these genes contained an LexA box, implicated in regulation of gene expression mediated by the SOS system, and the different positions of the LexA box could be partly responsible for the differences observed in terms of induction (89). The smaqnr and qnrD LexA-binding sites are found in both cases downstream of the 210 box sequence, in a similar position compared with qnrB1. Moreover, fluoroquinolones, as well as other antimicrobial agents, causing induction of the qnrB1, qnrD and smaqnr promoters, were regulated by the SOS system, in an RecA-dependent pathway, which was investigated successfully in 2012 (91).
Besides that, SOS is also known to promote HGT, which plays an essential role, especially for the antibiotic resistance development and dissemination among bacteria (1,92,93). The conjugative transfer of plasmids have been demonstrated to trigger a bacterial stress response—the SOS response—in recipient cells and can impact the cassette content of integrins (1). Integrating conjugative elements (ICEs), a diverse group of mobile elements that could recruit the SOS response to mobilize themselves from the bacterial chromosome and infect other cells, which transfers resistance to multiple antibiotics (92). Also, activation of the SOS response in both E. coli and V. cholerae greatly stimulates the transfer of SXT (a 100-kilobase ICE) and SXT-related elements (93). Another report on qnrVC3, which encodes a PRP of the Qnr subfamily, is present within a member of the SXT ICE family found commonly on the chromosomes of multidrug-resistant strains of V. cholerae and on the chromosomes of Escherichia coli transconjugants, proved to be accounted for transferable multidrug resistance that includes ciprofloxacin in isolates positive for qnrVC3 (94). Thus, the use of fluoroquinolones or some other antimicrobial agents, either clinically or in agricultural settings, causing induction of the SOS response, might potentiate the horizontal dissemination of antibiotic resistance genes to a broad range of bacterial species, and SOS response could then be a suitable target for co-treatment of infections in order to prevent exchange of antibiotic resistance/adaptation genes.
Conclusions
Hence, widespread use of fluoroquinolones has inevitably led to a sharp increase in the rate of resistance among different bacterial species in areas around the world, which the SOS response plays an unsuspected role and deserves comprehensive attention. As described above, we have a preliminary understanding of the induction, molecular mechanism, and modulation of fluoroquinolone resistance so as to search for effective ways to suppress the SOS network to reduce the number of resistant bacteria that arise from antibiotic treatment, and remains to be improved.
Acknowledgements
Funding: This work was supported by grant from the National Natural Science Foundation of China (81471994).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Baharoglu Z, Bikard D, Mazel D. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS Genet 2010;6:e1001165. [PubMed]
- Baharoglu Z, Mazel D. SOS, the formidable strategy of bacteria against aggressions. FEMS Microbiol Rev 2014;38:1126-45. [PubMed]
- Al Mamun AA, Lombardo MJ, Shee C, et al. Identity and function of a large gene network underlying mutagenic repair of DNA breaks. Science 2012;338:1344-8. [PubMed]
- Janion C. Inducible SOS response system of DNA repair and mutagenesis in Escherichia coli. Int J Biol Sci 2008;4:338-44. [PubMed]
- Erill I, Campoy S, Barbé J. Aeons of distress: an evolutionary perspective on the bacterial SOS response. FEMS Microbiol Rev 2007;31:637-56. [PubMed]
- Kohanski MA, Dwyer DJ, Hayete B, et al. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007;130:797-810. [PubMed]
- Liu Y, Imlay JA. Cell death from antibiotics without the involvement of reactive oxygen species. Science 2013;339:1210-3. [PubMed]
- Keren I, Wu Y, Inocencio J, et al. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 2013;339:1213-6. [PubMed]
- Sutton MD, Smith BT, Godoy VG, et al. The SOS response: recent insights into umuDC-dependent mutagenesis and DNA damage tolerance. Annu Rev Genet 2000;34:479-97. [PubMed]
- Courcelle J, Khodursky A, Peter B, et al. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 2001;158:41-64. [PubMed]
- Au N, Kuester-Schoeck E, Mandava V, et al. Genetic composition of the Bacillus subtilis SOS system. J Bacteriol 2005;187:7655-66. [PubMed]
- Goranov AI, Kuester-Schoeck E, Wang JD, et al. Characterization of the global transcriptional responses to different types of DNA damage and disruption of replication in Bacillus subtilis. J Bacteriol 2006;188:5595-605. [PubMed]
- Cirz RT, Jones MB, Gingles NA, et al. Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J Bacteriol 2007;189:531-9. [PubMed]
- Michel B. After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol 2005;3:e255. [PubMed]
- Singh R, Ledesma KR, Chang KT, et al. Impact of recA on levofloxacin exposure-related resistance development. Antimicrob Agents Chemother 2010;54:4262-8. [PubMed]
- Zhang AP, Pigli YZ, Rice PA. Structure of the LexA-DNA complex and implications for SOS box measurement. Nature 2010;466:883-6. [PubMed]
- Kovačič L, Paulič N, Leonardi A, et al. Structural insight into LexA-RecA* interaction. Nucleic Acids Res 2013;41:9901-10. [PubMed]
- Little JW. Mechanism of specific LexA cleavage: autodigestion and the role of RecA coprotease. Biochimie 1991;73:411-21. [PubMed]
- Butala M, Zgur-Bertok D, Busby SJ. The bacterial LexA transcriptional repressor. Cell Mol Life Sci 2009;66:82-93. [PubMed]
- Thi TD, López E, Rodríguez-Rojas A, et al. Effect of recA inactivation on mutagenesis of Escherichia coli exposed to sublethal concentrations of antimicrobials. J Antimicrob Chemother 2011;66:531-8. [PubMed]
- Lenhart JS, Brandes ER, Schroeder JW, et al. RecO and RecR are necessary for RecA loading in response to DNA damage and replication fork stress. J Bacteriol 2014;196:2851-60. [PubMed]
- Arnold DA, Kowalczykowski SC. Facilitated loading of RecA protein is essential to recombination by RecBCD enzyme. J Biol Chem 2000;275:12261-5. [PubMed]
- Courcelle J, Wendel BM, Livingstone DD, et al. RecBCD is required to complete chromosomal replication: Implications for double-strand break frequencies and repair mechanisms. DNA Repair (Amst) 2015;32:86-95. [PubMed]
- Morimatsu K, Wu Y, Kowalczykowski SC. RecFOR proteins target RecA protein to a DNA gap with either DNA or RNA at the 5' terminus: implication for repair of stalled replication forks. J Biol Chem 2012;287:35621-30. [PubMed]
- Rand L, Hinds J, Springer B, et al. The majority of inducible DNA repair genes in Mycobacterium tuberculosis are induced independently of RecA. Mol Microbiol. 2003;50:1031-42. [PubMed]
- López E, Elez M, Matic I, et al. Antibiotic-mediated recombination: ciprofloxacin stimulates SOS-independent recombination of divergent sequences in Escherichia coli. Mol Microbiol 2007;64:83-93. [PubMed]
- Gu B, Cao Y, Pan S, et al. Comparison of the prevalence and changing resistance to nalidixic acid and ciprofloxacin of Shigella between Europe-America and Asia-Africa from 1998 to 2009. Int J Antimicrob Agents 2012;40:9-17. [PubMed]
- Pfaller MA, Beach ML, Gordon KA, et al. Comparative antimicrobial spectrum and activity of BMS284756 (T-3811; a desfluoroquinolone) tested against an international collection of staphylococci and enterococci, including in vitro test development and intermethod comparisons. J Chemother 2002;14:13-8. [PubMed]
- Jacoby GA. Mechanisms of resistance to quinolones. Clin Infect Dis 2005;41 Suppl 2:S120-6. [PubMed]
- Veldman K, Cavaco LM, Mevius D, et al. International collaborative study on the occurrence of plasmid-mediated quinolone resistance in Salmonella enteric and Escherichia coli isolated from animals, humans, food and the environment in 13 European countries. J Antimicrob Chemother 2011;66:1278-86. [PubMed]
- Bonomo RA, Szabo D. Mechanisms of multidrug resistance in Acinetobacter species and Pseudomonas aeruginosa. Clin Infect Dis 2006;43 Suppl 2:S49-56. [PubMed]
- García-Fernández A, Gallina S, Owczarek S, et al. Emergence of Ciprofloxacin-Resistant Salmonella enterica Serovar Typhi in Italy. PLoS One 2015;10:e0132065. [PubMed]
- Wasyl D, Hoszowski A, Zając M. Prevalence and characterisation of quinolone resistance mechanisms in Salmonella spp. Vet Microbiol 2014;171:307-14. [PubMed]
- Jeong HS, Kim JA, Shin JH, et al. Prevalence of plasmid-mediated quinolone resistance and mutations in the gyrase and topoisomerase IV genes in Salmonella isolated from 12 tertiary-care hospitals in Korea. Microb Drug Resist 2011;17:551-7. [PubMed]
- Tamang MD, Nam HM, Kim A, et al. Prevalence and mechanisms of quinolone resistance among selected nontyphoid Salmonella isolated from food animals and humans in Korea. Foodborne Pathog Dis 2011;8:1199-206. [PubMed]
- Kim KY, Park JH, Kwak HS, et al. Characterization of the quinolone resistance mechanism in foodborne Salmonella isolates with high nalidixic acid resistance. Int J Food Microbiol 2011;146:52-6. [PubMed]
- Gunell M, Webber MA, Kotilainen P, et al. Mechanisms of resistance in nontyphoidal Salmonella enterica strains exhibiting a nonclassical quinolone resistance phenotype. Antimicrob Agents Chemother 2009;53:3832-6. [PubMed]
- Nüesch-Inderbinen M, Abgottspon H, Sägesser G, et al. Antimicrobial susceptibility of travel-related Salmonella enterica serovar Typhi isolates detected in Switzerland (2002-2013) and molecular characterization of quinolone resistant isolates. BMC Infect Dis 2015;15:212. [PubMed]
- Tamang MD, Nam HM, Chae MH, et al. Prevalence of plasmid-mediated quinolone resistance determinants among Escherichia coli isolated from food animals in Korea. Foodborne Pathog Dis 2012;9:1057-63. [PubMed]
- Gibson JS, Cobbold RN, Kyaw-Tanner MT, et al. Fluoroquinolone resistance mechanisms in multidrug-resistant Escherichia coli isolated from extraintestinal infections in dogs. Vet Microbiol 2010;146:161-6. [PubMed]
- Betitra Y, Teresa V, Miguel V, et al. Determinants of quinolone resistance in Escherichia coli causing community-acquired urinary tract infection in Bejaia, Algeria. Asian Pac J Trop Med 2014;7:462-7. [PubMed]
- Varela AR, Macedo GN, Nunes OC, et al. Genetic characterization of fluoroquinolone resistant Escherichia coli from urban streams and municipal and hospital effluents. FEMS Microbiol Ecol 2015.91. [PubMed]
- Baudry-Simner PJ, Singh A, Karlowsky JA, et al. Canadian Antimicrobial Resistance Alliance. Mechanisms of reduced susceptibility to ciprofloxacin in Escherichia coli isolates from Canadian hospitals. Can J Infect Dis Med Microbiol 2012;23:e60-4. [PubMed]
- Zhao L, Zhang J, Zheng B, et al. Molecular epidemiology and genetic diversity of fluoroquinolone-resistant Escherichia coli isolates from patients with community-onset infections in 30 Chinese county hospitals. J Clin Microbiol 2015;53:766-70. [PubMed]
- Zayed AA, Essam TM, Hashem AG, et al. 'Supermutators' found amongst highly levofloxacin-resistant E. coli isolates: a rapid protocol for the detection of mutation sites. Emerg Microbes Infect 2015;4:e4. [PubMed]
- Nagasaka Y, Kimura K, Yamada K, et al. Genetic profiles of fluoroquinolone- nonsusceptible Klebsiella pneumoniae among cephalosporin-resistant K. pneumoniae. Microb Drug Resist 2015;21:224-33. [PubMed]
- Lin CJ, Siu LK, Ma L, et al. Molecular epidemiology of ciprofloxacin-resistant extended-spectrum β-lactamase-producing Klebsiella pneumoniae in Taiwan. Microb Drug Resist 2012;18:52-8. [PubMed]
- Piekarska K, Wołkowicz T, Zacharczuk K, et al. Co-existence of plasmid-mediated quinolone resistance determinants and mutations in gyrA and parC among fluoroquinolone-resistant clinical Enterobacteriaceae isolated in a tertiary hospital in Warsaw, Poland. Int J Antimicrob Agents 2015;45:238-43. [PubMed]
- Ferjani S, Saidani M, Amine FS, et al. Prevalence and characterization of plasmid-mediated quinolone resistance genes in extended-spectrum β-lactamase-producing Enterobacteriaceae in a Tunisian hospital. Microb Drug Resist 2015;21:158-66. [PubMed]
- Singh P, Jain A, Dixit P, et al. Prevalence of gyrA and B gene mutations in fluoroquinolone-resistant and-sensitive clinical isolates of Mycobacterium tuberculosis and their relationship with MIC of ofloxacin. J Antibiot (Tokyo) 2015;68:63-6. [PubMed]
- Willby M, Sikes RD, Malik S, et al. Correlation between GyrA substitutions and ofloxacin, levofloxacin, and moxifloxacin cross-resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 2015;59:5427-34. [PubMed]
- Kim JS, Kim JJ, Kim SJ, et al. Outbreak of ciprofloxacin-resistant shigella sonnei associated with travel to Vietnam, Republic of Korea. Emerg Infect Dis 2015;21:1247-50. [PubMed]
- Azmi IJ, Khajanchi BK, Akter F, et al. Fluoroquinolone resistance mechanisms of Shigella flexneri isolated in Bangladesh. PLoS One 2014;9:e102533. [PubMed]
- Wang YH, Chen CL, Hou JN, et al. Serotype distribution and resistance genes associated with macrolide and fluoroquinolone resistance in Streptococcus agalactiae isolates from a hospital in southern Taiwan. Biomed J 2015;38:215-20. [PubMed]
- Dabboussi F, Allouche S, Mallat H, et al. Prevalence of first-step mutants among levofloxacin-susceptible isolates of Streptococcus pneumoniae in north Lebanon. J Chemother 2013;25:328-31. [PubMed]
- Chen M, Guo Q, Wang Y, et al. Shifts in the antibiotic susceptibility, serogroups, and clonal complexes of neisseria meningitidis in shanghai, china: a time trend analysis of the pre-quinolone and quinolone Eras. PLoS Med 2015;12:e1001838; discussion e1001838. [PubMed]
- Yang X, Xing B, Liang C, et al. Prevalence and fluoroquinolone resistance of Pseudomonas aeruginosa in a hospital of South China. Int J Clin Exp Med 2015;8:1386-90. [PubMed]
- Guo L, Zhang J, Xu C, et al. Molecular characterization of fluoroquinolone resistance in Haemophilus parasuis isolated from pigs in South China. J Antimicrob Chemother 2011;66:539-42. [PubMed]
- Gu DX, Hu YJ, Zhou HW, et al. Substitutions of Ser83Leu in GyrA and Ser80Leu in ParC Associated with quinolone resistance in acinetobacter pittii. Microb Drug Resist 2015;21:345-51. [PubMed]
- Maleki MH, Jalilian FA, Khayat H, et al. Detection of highly ciprofloxacin resistance Acinetobacter baumannii isolated from patients with burn wound infections in presence and absence of efflux pump inhibitor. Maedica (Buchar) 2014;9:162-7. [PubMed]
- Kong LC, Gao D, Gao YH, et al. Fluoroquinolone resistance mechanism of clinical isolates and selected mutants of Pasteurella multocida from bovine respiratory disease in China. J Vet Med Sci 2014;76:1655-7. [PubMed]
- Martínez-Martínez L, Pascual A, Jacoby GA. Quinolone resistance from a transferable plasmid. Lancet 1998;351:797-9. [PubMed]
- Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol 2013;303:298-304. [PubMed]
- Robicsek A, Strahilevitz J, Jacoby GA, et al. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat Med 2006;12:83-8. [PubMed]
- Rodríguez-Martínez JM, Cano ME, Velasco C, et al. Plasmid-mediated quinolone resistance: an update. J Infect Chemother 2011;17:149-82. [PubMed]
- Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis 2006;6:629-40. [PubMed]
- Yang H, Duan G, Zhu J, et al. Prevalence and characterisation of plasmid-mediated quinolone resistance and mutations in the gyrase and topoisomerase IV genes among Shigella isolates from Henan, China, between 2001 and 2008. Int J Antimicrob Agents 2013;42:173-7. [PubMed]
- Xiong Z, Li J, Li T, et al. Prevalence of plasmid-mediated quinolone-resistance determinants in Shigella flexneri isolates from Anhui Province, China. J Antibiot (Tokyo) 2010;63:187-9. [PubMed]
- Zhang W, Luo Y, Li J, et al. Wide dissemination of multidrug-resistant Shigella isolates in China. J Antimicrob Chemother 2011;66:2527-35. [PubMed]
- Tariq A, Haque A, Ali A, et al. Molecular profiling of antimicrobial resistance and integron association of multidrug-resistant clinical isolates of Shigella species from Faisalabad, Pakistan. Can J Microbiol 2012;58:1047-54. [PubMed]
- Folster JP, Pecic G, Bowen A, et al. Decreased susceptibility to ciprofloxacin among Shigella isolates in the United States, 2006 to 2009. Antimicrob Agents Chemother 2011;55:1758-60. [PubMed]
- Bhattacharya D, Bhattacharjee H, Thamizhmani R, et al. Prevalence of the plasmid-mediated quinolone resistance determinants among clinical isolates of Shigella sp. in Andaman & Nicobar Islands, India. Lett Appl Microbiol 2011;53:247-51. [PubMed]
- Ceyssens PJ, Mattheus W, Vanhoof R, et al. Trends in serotype distribution and antimicrobial susceptibility in Salmonella enterica isolates from humans in Belgium, 2009 to 2013. Antimicrob Agents Chemother 2015;59:544-52. [PubMed]
- Colobatiu L, Tabaran A, Flonta M, et al. First description of plasmid-mediated quinolone resistance determinants and β-lactamase encoding genes in non-typhoidal Salmonella isolated from humans, one companion animal and food in Romania. Gut Pathog 2015;7:16. [PubMed]
- Yang J, Luo Y, Cui S, et al. Diverse phenotypic and genotypic characterization among clinical Klebsiella pneumoniae and Escherichia coli isolates carrying plasmid-mediated quinolone resistance determinants. Microb Drug Resist 2011;17:363-7. [PubMed]
- Li L, Wang B, Feng S, et al. Prevalence and characteristics of extended-spectrum β-lactamase and plasmid-mediated fluoroquinolone resistance genes in Escherichia coli isolated from chickens in Anhui province, China. PLoS One 2014;9:e104356. [PubMed]
- Chmielarczyk A, Pobiega M, de Champs C, et al. The high prevalence of plasmid-mediated quinolone resistance among very low birth-weight infants in Poland. Microb Drug Resist 2015;21:391-7. [PubMed]
- Jamborova I, Dolejska M, Vojtech J, et al. Plasmid-mediated resistance to cephalosporins and fluoroquinolones in various Escherichia coli sequence types isolated from rooks wintering in Europe. Appl Environ Microbiol 2015;81:648-57. [PubMed]
- Okade H, Nakagawa S, Sakagami T, et al. Characterization of plasmid-mediated quinolone resistance determinants in Klebsiella pneumoniae and Escherichia coli from Tokai, Japan. J Infect Chemother. 2014;20:778-783. [PubMed]
- Crémet L, Caroff N, Dauvergne S, et al. Prevalence of plasmid-mediated quinolone resistance determinants in ESBL Enterobacteriaceae clinical isolates over a 1-year period in a French hospital. Pathol Biol (Paris) 2011;59:151-6. [PubMed]
- Vervoort J, Gazin M, Kazma M, et al. High rates of intestinal colonisation with fluoroquinolone-resistant ESBL-harbouring Enterobacteriaceae in hospitalised patients with antibiotic-associated diarrhoea. Eur J Clin Microbiol Infect Dis 2014;33:2215-21. [PubMed]
- Poole K. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother 2005;56:20-51. [PubMed]
- Ruiz J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother 2003;51:1109-17. [PubMed]
- Cirz RT, Romesberg FE. Induction and inhibition of ciprofloxacin resistance-conferring mutations in hypermutator bacteria. Antimicrob Agents Chemother 2006;50:220-5. [PubMed]
- Dörr T, Lewis K, Vulić M. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genet 2009;5:e1000760. [PubMed]
- Schröder W, Goerke C, Wolz C. Opposing effects of aminocoumarins and fluoroquinolones on the SOS response and adaptability in Staphylococcus aureus. J Antimicrob Chemother 2013;68:529-38. [PubMed]
- Urios A, Herrera G, Aleixandre V, et al. Influence of recA mutations on gyrA dependent quinolone resistance. Biochimie 1991;73:519-21. [PubMed]
- Yim G, McClure J, Surette MG, et al. Modulation of Salmonella gene expression by subinhibitory concentrations of quinolones. J Antibiot (Tokyo) 2011;64:73-8. [PubMed]
- Da Re S, Garnier F, Guérin E, et al. The SOS response promotes qnrB quinolone-resistance determinant expression. EMBO Rep 2009;10:929-33. [PubMed]
- Wang M, Jacoby GA, Mills DM, et al. SOS regulation of qnrB expression. Antimicrob Agents Chemother 2009;53:821-3. [PubMed]
- Briales A, Rodriguez-Martinez JM, Velasco C, et al. Exposure to diverse antimicrobials induces the expression of qnrB1, qnrD and smaqnr genes by SOS-dependent regulation. J Antimicrob Chemother 2012;67:2854-9. [PubMed]
- Hastings PJ, Rosenberg SM, Slack A. Antibiotic-induced lateral transfer of antibiotic resistance. Trends Microbiol 2004;12:401-4. [PubMed]
- Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 2004;427:72-4. [PubMed]
- Kim HB, Wang M, Ahmed S, et al. Transferable quinolone resistance in Vibrio cholerae. Antimicrob Agents Chemother 2010;54:799-803. [PubMed]


