Beyond the World Health Organization grading of infiltrating gliomas: advances in the molecular genetics of glioma classification
Introduction
Diffuse infiltrating gliomas (DIGs) are the second most common primary central nervous system (CNS) neoplasm and account for 80% of primary malignant gliomas. The total incidence of primary CNS tumors is approximately 18.7 per 100,000 persons in the United States and 7 per 100,000 worldwide. While primary CNS tumors account for only 2% of primary tumors, they cause 7% of the years of life lost from cancer before age 70. More than half of these gliomas are glioblastomas (GBMs) for which the overall 5-year survival rate remains less than 5% (1-6).
Current glioma classifications are based on the 2007 World Health Organization (WHO) grading scale, which separates gliomas based on cytologic features and degrees of malignancy after hematoxylin and eosin (H&E) staining (Table 1). This system was first employed in the 1920s when Bailey and Cushing first classified glial tumors by their similarity to known glial cell types: astrocytes, oligodendrocytes, etc. Thus the cytologic features place diffuse gliomas into large categories of astrocytomas, oligodendrogliomas, and mixed oligoastrocytomas (7,8). Infiltrating gliomas are graded as WHO II-IV (Grade I are typically solid and non-infiltrative tumors such as pilocytic astrocytomas and subependymal giant cell astrocytomas); histologic grading is based on findings of nuclear atypia, proliferative (mitotic) activity, microvascular proliferation, and necrosis (9). Currently, the WHO classification scheme remains a practical and effective means to classify gliomas and is the only widely accepted system. However, this system is based solely on histologic visual criteria and prone to subjective inter-observer variation. Many cases fail to adhere to one class and are not reliably associated with tumor aggressiveness, treatment response, and ultimately progression-free survival (8). Interpreting cellularity, anaplasia, or even cell type is not always possible due to the histological heterogeneity of tumors (7). Tumors of the same WHO grade can have a different clinical course. One solution to this conundrum is to use molecular and cytogenetic information to assist in diffuse glioma classification. In concert with histologic assessment, genetic sub-grouping of gliomas may produce more accurate and reproducible diagnostic criteria than the current system. Hence, treatment protocols could be established based on genetic background and copy number abnormalities (CNAs). Improving glioma classification may impact patient treatment and survival.
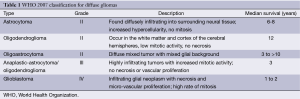
Full table
In this review, we aim to use the existing WHO classification of diffuse gliomas (grade II-IV) and incorporate the most recent advances in genetic and molecular subclassification schemes within each group. Beginning with a brief description of four criteria used to help with subtyping gliomas, we will then apply these groupings to the WHO classifications (Table 2). The aim of translational medicine remains to take this new wealth of information and implant it within the framework of our existing clinical schema to enhance clinical decision making, and ultimately improve patient treatments and prognoses.
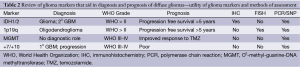
Full table
Isocitrate dehydrogenase mutations
The classification of infiltrating gliomas in adults has been altered by the discovery and characterization of the IDH1 and IDH2 mutations in a subset of these neoplasms. Molecular profiling based on IDH mutational status has been described over the last 6 years as a significant prognostic marker; further investigation has clarified that IDH mutant tumors constitute a genetically and clinically distinct group of neoplasms as compared to their IDH wild-type counterparts (10).
IDH1 mutations were first described in GBM (WHO grade IV) tumors. Continued research has focused on the role of IDH mutations in lower grade gliomas (11). IDH mutations are found in a minority (12%) of GBMs but in approximately 70% of grade II-III infiltrating gliomas (9,12). IDH mutations are subdivided into IDH1 vs. IDH2 mutations. Approximately 90% of IDH mutations occur with IDH1. The most common IDH1 mutation is a R132H point mutation. Several other mutations are described in the literature at codon 132 and codon 172 for IDH2 (9).
Research into CNS tumorigenesis has found that IDH mutations take part in the initial transformation of a glial cell into a tumor cell; proposed models of gliomagenesis suggest that the early event of an IDH mutation is followed by further changes along stereotypic molecular and cytogenetic pathways corresponding to “oligodendroglial” or “astrocytic” differentiation (13). Broadly, molecular profiling studies have noted that most neoplasms with an “oligodendroglial” profile as well as a subset of neoplasms with an “astrocytic” profile carry IDH1/2 mutations (14).
Overall, IDH1/2 mutations serve as important biomarkers for diffuse gliomas. IDH mutant gliomas behave less aggressively and have a better prognosis as compared to IDH wild-type gliomas. The finding of an IDH mutation confers a better prognosis regardless of tumor grade and other variables, serving as an independent prognostic factor (11,15). Though IDH mutations can be the precursor to CNS glioma formation, the presence of IDH mutations is associated with a positive predictive value for better progression free survival and overall survival (16).
Whether mutations in IDH result in a loss of tumor suppressor function, or act as an oncogene, remains a source of debate. It has been hypothesized that the effects of the IDH mutant enzyme’s product, 2-hydroxyglutarate (2-HG), changes a cell’s methylation profile, alters cell telomere length, and gene expression. The effects of IDH mutations also produce decreased cytoplasmic levels of alpha-ketoglutarate and NAPDH that may in turn stabilize hypoxia inducible factor 1-α facilitating cellular proliferation (17,18).
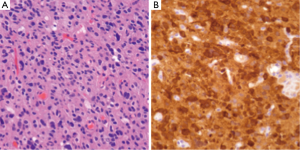
IDH1/2 mutations can be assessed through several methods. As these are point mutations, several polymerase chain reaction (PCR) assays are available. Alternatively, immunohistochemistry (IHC) for IDH1 mutant protein can be employed to study tissue samples (Figure 1) by testing for the R132H mutation. Assays for IDH1 mutations can not only serve as methods to provide prognostic information regarding known neoplasms, but also can aid in the original diagnosis of neoplasia (19).
1p/19q co-deletion
There is great inter-observer variability in classifying gliomas as pure astrocytomas, oligodendrogliomas, or mixed oligoastrocytomas leading to diagnostic uncertainty and varying treatment practices. Unbalanced translocation of chromosomes 1 and 19 with deletion of the 1p arm and the 19q arm is found in close to 70% of histologically defined oligodendrogliomas. It is reported this number may be as high as 90% using strict histological criteria (14,20,21).
A 1p/19q codeletion is considered to be the objective molecular definition of oligodendroglial lineage, and tumors that lack the codeletion are often observed to have ATRX and TP53 mutations, which are putative markers for astrocytic lineage. Further, 1p/19q codeletions and ATRX/TP53 mutations are relatively exclusive of one another, supporting the argument of two separate lineages that can be identified on a molecular basis to supplement histological diagnosis. The 1p/19q loss may be itself a mechanism of inactivation of CIC and FUBP1. Profiling studies show that FUBP1 and CIC mutations occur simultaneously with IDH mutations, creating a unique molecular profile for oligodendrogliomas (22).
Oligodendrogliomas with a 1p/19q co-deletion behave in an indolent fashion and tend to respond well to chemotherapy (20). Testing for the presence of a 1p/19q co-deletion is essential for gliomas that appear oligodendroglial in origin. FISH analysis, multiplex PCR, and SNP array are common techniques used to test for the co-deletion. A 1p19q codeletion is associated with improved prognosis in low-grade gliomas (LGGs) and predictive of improved outcomes with chemotherapy and radiation (14,21,23).
O6-methyl-guanine-DNA methyltransferase (MGMT) status
The DNA repair enzyme MGMT repairs O6 alkyl guanine adducts. The repair enzyme mechanism interferes with the overlapping effect of temozolamide (TMZ), which alkylates at the O6 position of guanine. The MGMT gene has a 5'-promoter region that contains a CpG island, and methylation of the CpG island results in epigenetic silencing of gene transcription (23,24). MGMT promoter methylation was identified in 36% of the overall population of glioma series studied which demonstrated the presence of the mutation improves the effect of TMZ and ultimately increases overall survival regardless of treatment arm (25). Numerous trials have shown that MGMT promoter methylation is a prognostic marker associated with improved survival (25-28). MGMT promoter methylation can be associated with 1p19q codeletions as well as IDH mutations suggesting it may be an epiphenomenon related to these or other factors that result in improved survival (29).
Genetic classifications
The recent trend towards genomic profiling of gliomas has led to the exploration of classifying lesions into molecular groups based on multi-gene predictors. This classification may allow for the identification of new targets with the additional benefits of predictive markers for target therapies. The Cancer Genome Atlas (TCGA) database first sequenced GBM in 2008. Researchers have examined DNA, mRNA, microRNA and epigenetic profiling to identify multiple glioma subtypes with different clinical outcomes. Genetic abnormalities involving gain and loss of heterozygosity (LOH) of chromosome 7 and 10 respectively with associated EGFR amplification and PTEN loss drive tumorigenesis in high-grade gliomas (30,31). Similarly, the presence of mutations involved in the receptor tyrosine kinase (RTK), p53, or RB pathways have similar associations with high-grade gliomas. Ultimately, patient specific data from high throughput screening will be utilized for diagnosis, prognostication, and patient-specific therapy based on the unique genetic signature of their neoplasm (Figures 2,3).

Gliomas
Low grade gliomas (LGGs): WHO grade II
Epidemiology
Low-grade infiltrating gliomas (LGG; WHO grade II) account for 28% of primary CNS tumors in the United States and often progress to high-grade gliomas (WHO grade III and IV). Oligodendrogliomas make up 5% and astrocytomas make up 17.4% with the remaining being classified as oligoastrocytomas. Patients with grade II lesions have a 5-year survival rate of approximately 50% without taking into account any molecular subclassifications (32). The median age at time of diagnosis ranges between 43 and 48, depending on histologic subtype (33).
Pathology/IHC
Basic histological classification of a LGG is based on cytologic evaluation of astrocytic or oligodendroglial differentiation. Based on the 2007 WHO classification, astrocytic and oligodendroglial gliomas are divided into three histological types: diffuse astrocytoma, oligoastrocytomas, and oligodendrogliomas (9). In all types there is diffuse invasion without a clear tumor border in addition to nuclear atypia as compared to non-neoplastic cellular counterparts. Individual tumor cells may show nuclear irregularity (in the case of astrocytomas) or round nuclei with perinuclear clearing (in the case of oligodendrogliomas). Some tumors show a mix of cellular morphologies, leading to the designation of oligoastrocytoma (9).
Clinically, most diffuse astrocytomas are well-differentiated and slow growing, with a tendency to recur after surgical resection, often with progression to higher grade gliomas (8). Oligodendrogliomas are typically even more indolent; however, these lesions also eventually progress to higher grade lesions (34).
Immunohistochemical staining for glial fibrillary acidic protein (GFAP) often permits confirmation of glial origin. Immunohistochemical staining for mitotic activity using the antibody MIB-1 can also be employed to verify a low proliferative index (35). Additional staining for p53 can assist in the designation of a lesion as a neoplastic glial process and is more commonly positive in astrocytic tumors (36). Currently, these criteria are employed to make the final pathological diagnosis of a WHO II glioma. Further stratification based on molecular and genetic changes must be considered to enhance a clinician’s ability to determine prognosis and treatment course.
Genetics and molecular markers
A review of literature over the last five years indicates that there is a growing consensus that patients diagnosed with LGG should be stratified into two groups based on 1p/19q codeletion and IDH mutational status (17,20,37). Genetically, LGGs contain IDH1 mutations in 70-80% of cases. Additionally TP53 mutations are seen in ~60% of astrocytomas whereas over 70% of histologically defined oligodendrogliomas show codeletion of 1p/19q. The mixed oligoastrocytomas carry either TP53 mutations or 1p/19q co-deletions as these alterations are for the most part mutually exclusive (38,39). IDH mutations and 1p/19q co-deletion have been studied extensively in retrospective and prospective trials and are shown to be associated with improved prognosis.
The workup of infiltrating LGGs in an adult requires determination of its nature as an astrocytic or oligodendroglial neoplasm. Aside from histological findings as discussed above, testing for molecular and cytogenetic features can aid in classification and provides additional information for patients and treating clinicians. Most important is the definition of a tumor’s IDH mutation status, which can be assessed through IHC (for the protein product of the most common mutation) or molecular methods (19). Second, delineation of a 1p/19q co-deletion within an infiltrating glioma provides cytogenetic evidence of an oligodendroglial tumor and places the tumor in a distinct category with prognostic and predictive relevance (40). Third, identification of an ATRX or TP53 mutation supports the interpretation that the neoplasm is an infiltrating astrocytoma, since these abnormalities are rarely seen in oligodendrogliomas (Figures 4,5) (38,39). Molecular profiling has shown that infiltrating astrocytomas in adults with ATRX and TP53 mutations often co-segregate with IDH mutations and behave in a more indolent fashion than IDH wild-type astrocytomas (14,21,39). Use of PCR assays or next-generation sequencing in addition to IHC can help identify these mutations (Figures 2,3) (35,38).
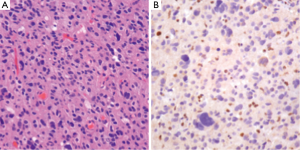
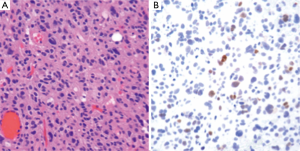
Ultimately, one can use this data to stratify three groups of patients with histological LGGs: 1p/19q co-deletion with IDH mutations and 1p/19q intact patients with or without IDH mutations (1p/19q co-deletion is almost always associated with IDH mutations) (Table 3). Those patients with 1p/19q co-deletions are diagnosed with oligodendrogliomas and it is believed that this codeletion will eventually become a requirement for the diagnosis of oligodendrogliomas. In combination with 1p/19q assessment, use of TP53 and ATRX evaluation may allow for definitive molecular classification of tumors thought to be mixed by histologic parameters (oligoastrocytomas) (39). Research continues into identifying other molecular markers that may help with prognostication in LGGs.

Full table
In clinical practice, oligodendroglioma patients, without chromosomal 1p19q codeletions, may need to be followed more closely with serial imaging at shorter time intervals. Patients with WHO grade II astrocytomas, without IDH mutations, are prone to transform to higher grade tumors faster than those patients with IDH mutations present in their tumor. These patients tend to have a poorer prognosis (6).
Anaplastic astrocytomas/oligodendrogliomas (AA/AOs): WHO grade III
Epidemiology
Anaplastic malignant gliomas (WHO grade III) exhibit nuclear pleomorphism and most importantly increased mitotic activity (9). They are a heterogeneous group classified as AAs, anaplastic oligoastrocytomas (AOA), and AOs. These tumors account for 6.7% of gliomas and patients have a 5-year survival of approximately 30% with the majority of patients progressing to grade IV GBM (32). The accurate histological diagnosis of anaplastic gliomas is of great importance for determining a patient’s prognosis and guiding therapy.
Pathology/IHC
The WHO classification of CNS tumors as AAs, oligodendrogliomas, or oligoastrocytomas is based on similar architectural and cytologic features as each tumor’s grade II counterpart. However, anaplastic infiltrating gliomas are distinguished from lower grade lesions primarily by the presence of mitotic activity. Anaplastic gliomas additionally often show greater degrees of cellularity and nuclear atypia than their grade II counterparts, but mitotic activity serves as the primary discriminant between grade II and grade III lesions. AO may also feature tumor necrosis and vascular proliferation in addition to elevated proliferation indices; under current criteria pure oligodendroglial tumors are not classified as grade IV neoplasms (GBM) without a definable astrocytic component (9).
Pathologic evaluation of these lesions is similar to lower grade gliomas, with the added caveat of close inspection for focal necrosis or vascular proliferation that may indicate a higher grade process. As is true for lower grade lesions, a typical immunohistochemical staining panel may include IDH1 mutant protein, p53, ATRX and MIB-1 for proliferation index (22,39,41). AOA, like their grade II counterparts, may be best addressed for definitive classification as either astrocytoma or oligodendroglioma by molecular means (38,39).
Genetics and molecular markers
AAs carry a worse prognosis than AOs. AAs progress from WHO II astrocytomas and typically harbor the same genetic precursor mutations of IDH1/2, TP53 and ATRX. Early concomitant mutations in these genes lead to the formation of LGGs that progress to higher grade anaplastic lesions. Stratification by IDH status is key. IDH wild-type gliomas tend to be higher grade than IDH mutant gliomas and also tend to be more aggressive when matched grade-for-grade with their IDH mutant counterparts. Particularly when higher grade (WHO grade III to IV), IDH wild-type gliomas often acquire certain cytogenetic abnormalities, namely amplifications of EGFR and deletions of PTEN (42,43).
Malignant progression has been associated both with particular genetic abnormalities and with an increase in the number of aberrations; in fact, IDH wild-type anaplastic astrocytomas often show cytogenetic abnormalities similar to those of glioblastoma (11). The most common copy number aberrations identified in GBM exist on chromosomes 7 and 10, where EGFR and PTEN are located. Amplifications of EGFR are strongly associated with poor prognosis and progression to WHO IV lesions. EGFR amplifications are seen in 30-40% of GBMs and appear to be by and large mutually exclusive with IDH mutations in high-grade gliomas. PTEN deletions are seen in a much higher proportion (approximately 80%) of GBMs. The finding of one or both of these abnormalities reinforces the diagnosis, tumor grade, and classification as an IDH wild-type tumor. Stratification of astrocytomas based on copy number aberrations and the accumulation of mutations on chromosomes 7, 9, and 10 will ultimately serve as predictors of outcome (1) (Table 4).

Full table
As discussed with WHO grade II oligodendrogliomas, AOs with 1p/19q co-deletion have been associated with increased chemosensitivity and longer progression free survival (22). Studies of comparative genomic hybridization (CGH) data on AOs show many features in common with well-differentiated oligodendrogliomas with additional deviations of gains on chromosome 7 and losses on 4, 9p, and 10. Additionally, recurrent oligodendrogliomas, which have progressed to WHO grade III, have similar gains on 7 and losses on 10. These gains of 7 and losses of 10 are associated with EGFR amplification and PTEN loss seen similarly in GBM and associated with equally poor prognosis (14,20,44). Those tumors with 1p/19q loss or 7q gain also show close correlation with IDH mutations, which are consistent with their overall improved prognosis. Even though oligodendrogliomas are known to have improved prognosis, IDH1 mutants have improved progression free survival compared to IDH1 wild type cases (5,45).
In clinical practice, patients with anaplastic (WHO grade III) gliomas will be followed with short interval imaging and clinic visits. Those patients with tumors containing MGMT methylation and IDH mutations will have a better prognosis than those patients who are MGMT unmethylated with wild-type IDH status (12,20,24,37,41). High-grade gliomas that harbor 1p19q co-deletions also have improved prognosis (22). The accumulation of cytogenetic abnormalities and particularly gain of chromosome 7 and loss of chromosome 10 are indicators of likely transformation to WHO grade IV tumors, also known as GBM.
Glioblastoma (GBM): WHO IV
Epidemiology
The most common adult glioma is a GBM (WHO grade IV astrocytoma) accounting for 15% of primary brain and CNS tumors and 55% of all gliomas. The incidence of GBM is 3.19/100,000 with a 5-year survival rate for patients <5%. GBM is the most aggressive diffuse glioma of the astrocytic lineage with patient median survival of ranging from 12-15 months despite all therapies.
Pathology/IHC
Under current classification, GBMs are infiltrating astrocytomas that share the enhanced mitotic activity of anaplastic lesions and carry additional histologic features of vascular proliferation and tumor necrosis. Usually, GBMs are highly cellular tumors with marked nuclear atypia and “pseudo-palisading” necrosis characterized by the heaping up of tumor cells surrounding zones of necrosis. Oftentimes thrombi are noted within vessels inside and adjacent to the tumor; these thrombi may also be associated with necrotic foci (9). Complete microscopic resection can never be achieved and recurrence is common following therapy (9). There are a variety of histological variants of GBM that include small cell, giant cell, and gliosarcoma. Some histologic subtypes of GBM may introduce diagnostic challenges that require immunohistochemical confirmation of the tumor’s glial nature; GFAP immunostaining is usually helpful in these scenarios. Further immunohistochemical workup of GBMs may involve staining for IDH1 mutant protein, ATRX, and p53, similar to lower grade astrocytomas (19,38).
Genetics/molecular markers
All GBMs can be divided into two subtypes based on the presence of absence of a precursor lesion of a lower grade. Primary GBM is the most common type (>90%) and diagnosed as a de novo lesion without progression from a lower grade tumor in older patients (>60 years). Secondary GBMs result from progression of LGGs (WHO grade II/III) and are commonly found as a recurrence in younger patients. The time to GBM progression from a grade II in contrast to a grade III lesion is also longer (5 to 2 years respectively) (13,46,47). Progression to a GBM can occur at any time though. The identification of GBMs as primary vs. secondary was further sub-classified after the analysis of the TGCA data derived from GBM patients. Over the last 5 years, numerous research groups have analyzed the data to generate defined subclasses of GBM based on genetic and molecular profiles that correlate to prognosis and treatment response.
Analysis of GBM genomics reveals multiple tumor suppressor and oncogenes that are inactive and active, respectively, during tumor progression and de novo formation. The three main pathways implicated in GBM formation are: RTK-RAS-MAPK-PI3KA, the p53 pathway, and the RB pathway (4,6,23,48,49). As discussed in the prior sections, the gain of copies of chromosome 7 along with losses of chromosome 10 contributes to the EGFR and PTEN mutations commonly identified in primary GBMs. IDH mutations, TP53 mutations, and ATRX mutations are characteristic of secondary GBMs (21,39). These mutations cluster specifically into gene expression profiles that are characteristic of recently described GBM subgroups.
TCGA is a global genomic profiling project that utilized high-throughput microarray technologies to identify molecular subtype classifications of cancers, multigene clinical predictors, new targets for drug therapy, and predictive markers for these therapies. In the case of GBM, two major studies of the TCGA data identified three major subtypes (50,51). Two of the subgroups consistently replicate similar profiles in various studies and are in stark contrast to each other: proneural and mesenchymal. Proneural GBM is a secondary GBM that is present in young adults and has neuronal differentiation that is associated with better outcomes. Proneural GBMs have IDH and TP53 mutations, glioma-CpG island methylator phenotype, and normal expression of EGFR/PTEN. This group represents close to 10% of all GBMs, consistent with the known prevalence of secondary GBMs (6,50-52). The mesenchymal GBM is common in older adults and is associated with a worse prognosis and characterized by NF1 loss or mutations, abnormalities in Akt signaling, increased expression of angiogenic peptides, and overexpression of genes related to motility, the extracellular matrix, and cell adhesion. The final TCGA subtypes are the neural and classical tumors that are associated with PTEN loss and EGFR amplification and constitute the majority of GBMs (Figure 6).
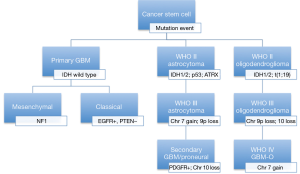
Sequence-based analyses of the TCGA data missed unique fusion mutations with oncogenic potential. Singh et al. first described chromosomal translocations that fuse in-frame the tyrosine kinase coding domains of fibroblast growth factor receptor (FGFR) genes (FGFR1 or FGFR3) to the transforming acidic coiled-coil (TACC) coding domains of TACC1 or TACC3, respectively. The FGFR-TACC fusion protein displays oncogenic activity when introduced into astrocytes causing mitotic defects leading to aneuploidy and ultimately gliomagenesis (53). The first studies found close to 5-7% of examined GBMs harbored this unique fusion protein. Tumorigenesis in this subset of GBMs is thought to be initiated by this fusion protein with its growth promoting function and loss of mitotic control leading to aneuploidy and ultimately tumor progression (53). As a tumor initiator, the fusion protein is a favorable target for therapy. This fusion protein’s oncogenic activity is achieved by constitutively activated FGFR that has become the target of kinase inhibitors in drug trials (54).
Recently, two studies have associated mutations in the promoter region of the telomerase reverse transcriptase (TERTp) with worse prognosis in GBM. This finding was predicated on prior studies showing increased telomerase activity has been associated with poor-prognosis in high grade gliomas. In a study of over 395 GBM biopsy samples, two TERTp mutations were present in over 75% of the samples and associated with poor survival (55). A second group studied 192 samples and found a similar link between TERTp mutation status and survival establishing a link between the mutation and an aggressive clinical course if not treated with surgical resection and chemotherapy (56). Ultimately, the TERTp mutations can be added to the list of prognostic biomarkers, including the aforementioned EGFR and IDH1 mutational status that will further aid in stratification of patient prognosis and ultimately therapy (57). The importance in identifying these subtypes lies in the enrichment of specific mutations for each subtype that could be potential therapeutic targets for personalized clinical trials.
Discussion
The use of the WHO grading scale will remain a cornerstone in the diagnosis, treatment, and prognostication of diffuse gliomas. However, the advance in molecular profiling and identification of unique gene expression profiles, key mutations, fusion proteins, and cytogenetic events can separate diffuse gliomas into subtypes that lend themselves to personalized therapies and prognostic groups. Using these tools will better account for clinical, pathologic, and molecular heterogeneity observed in WHO tumor grades. Doing so will allow clinicians to ultimately design better clinical trials specific to glioma tumor types and identify unique therapeutic targets based on the individual patient profile.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Hirose Y, Sasaki H, Abe M, et al. Subgrouping of gliomas on the basis of genetic profiles. Brain Tumor Pathol 2013;30:203-8. [PubMed]
- Kim YW, Koul D, Kim SH, et al. Identification of prognostic gene signatures of glioblastoma: a study based on TCGA data analysis. Neuro Oncol 2013;15:829-39. [PubMed]
- Olar A, Aldape KD. Biomarkers classification and therapeutic decision-making for malignant gliomas. Curr Treat Options Oncol 2012;13:417-36. [PubMed]
- Olar A, Aldape KD. Using the molecular classification of glioblastoma to inform personalized treatment. J Pathol 2014;232:165-77. [PubMed]
- Sulman EP, Guerrero M, Aldape K. Beyond grade: molecular pathology of malignant gliomas. Semin Radiat Oncol 2009;19:142-9. [PubMed]
- Theeler BJ, Yung WK, Fuller GN, et al. Moving toward molecular classification of diffuse gliomas in adults. Neurology 2012;79:1917-26. [PubMed]
- Erridge SC, Hart MG, Kerr GR, et al. Trends in classification, referral and treatment and the effect on outcome of patients with glioma: a 20 year cohort. J Neurooncol 2011;104:789-800. [PubMed]
- Pollo B. Neuropathological diagnosis of brain tumours. Neurol Sci 2011;32 Suppl 2:S209-11. [PubMed]
- Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 2007;114:97-109. [PubMed]
- Appin CL, Brat DJ. Molecular genetics of gliomas. Cancer J 2014;20:66-72. [PubMed]
- Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med 2009;360:765-73. [PubMed]
- Kurian KM, Haynes HR, Crosby C, et al. IDH mutation analysis in gliomas as a diagnostic and prognostic biomarker. Br J Neurosurg 2013;27:442-5. [PubMed]
- Huse JT, Wallace M, Aldape KD, et al. Where are we now? And where are we going? A report from the Accelerate Brain Cancer Cure (ABC2) low-grade glioma research workshop. Neuro Oncol 2014;16:173-8. [PubMed]
- Suzuki A, Nobusawa S, Natsume A, et al. Olig2 labeling index is correlated with histological and molecular classifications in low-grade diffuse gliomas. J Neurooncol 2014;120:283-91. [PubMed]
- Lin N, Yan W, Gao K, et al. Prevalence and clinicopathologic characteristics of the molecular subtypes in malignant glioma: a multi-institutional analysis of 941 cases. PLoS One 2014;9:e94871. [PubMed]
- Zou P, Xu H, Chen P, et al. IDH1/IDH2 mutations define the prognosis and molecular profiles of patients with gliomas: a meta-analysis. PLoS One 2013;8:e68782. [PubMed]
- Kloosterhof NK, Bralten LB, Dubbink HJ, et al. Isocitrate dehydrogenase-1 mutations: a fundamentally new understanding of diffuse glioma? Lancet Oncol 2011;12:83-91. [PubMed]
- Thompson CB. Metabolic enzymes as oncogenes or tumor suppressors. N Engl J Med 2009;360:813-5. [PubMed]
- Agarwal S, Sharma MC, Jha P, et al. Comparative study of IDH1 mutations in gliomas by immunohistochemistry and DNA sequencing. Neuro Oncol 2013;15:718-26. [PubMed]
- Boots-Sprenger SH, Sijben A, Rijntjes J, et al. Significance of complete 1p/19q co-deletion, IDH1 mutation and MGMT promoter methylation in gliomas: use with caution. Mod Pathol 2013;26:922-9. [PubMed]
- Weller M, Pfister SM, Wick W, et al. Molecular neuro-oncology in clinical practice: a new horizon. Lancet Oncol 2013;14:e370-9. [PubMed]
- Jiang H, Ren X, Cui X, et al. 1p/19q codeletion and IDH1/2 mutation identified a subtype of anaplastic oligoastrocytomas with prognosis as favorable as anaplastic oligodendrogliomas. Neuro Oncol 2013;15:775-82. [PubMed]
- Masui K, Cloughesy TF, Mischel PS. Review: molecular pathology in adult high-grade gliomas: from molecular diagnostics to target therapies. Neuropathol Appl Neurobiol 2012;38:271-91. [PubMed]
- Leu S, von Felten S, Frank S, et al. IDH/MGMT-driven molecular classification of low-grade glioma is a strong predictor for long-term survival. Neuro Oncol 2013;15:469-79. [PubMed]
- Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 2005;352:997-1003. [PubMed]
- Bady P, Sciuscio D, Diserens AC, et al. MGMT methylation analysis of glioblastoma on the Infinium methylation BeadChip identifies two distinct CpG regions associated with gene silencing and outcome, yielding a prediction model for comparisons across datasets, tumor grades, and CIMP-status. Acta Neuropathol 2012;124:547-60. [PubMed]
- Weller M, Stupp R, Reifenberger G, et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol 2010;6:39-51. [PubMed]
- Wick W, Weller M, van den Bent M, et al. MGMT testing--the challenges for biomarker-based glioma treatment. Nat Rev Neurol 2014;10:372-85. [PubMed]
- Sahebjam S, McNamara MG, Mason WP. Emerging biomarkers in anaplastic oligodendroglioma: implications for clinical investigation and patient management. CNS Oncol 2013;2:351-8. [PubMed]
- Kalinina J, Peng J, Ritchie JC, et al. Proteomics of gliomas: initial biomarker discovery and evolution of technology. Neuro Oncol 2011;13:926-42. [PubMed]
- Van Meir EG, Hadjipanayis CG, Norden AD, et al. Exciting new advances in neuro-oncology: the avenue to a cure for malignant glioma. CA Cancer J Clin 2010;60:166-93. [PubMed]
- Ostrom QT, Gittleman H, Farah P, et al. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006-2010. Neuro Oncol. 2013;15 Suppl 2:ii1-56. [PubMed]
- Ostrom QT, Gittleman H, Liao P. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2007–2011, Neuro Oncol 2014;16 suppl 4:iv1-63. [PubMed]
- El-Hateer H, Souhami L, Roberge D, et al. Low-grade oligodendroglioma: an indolent but incurable disease? Clinical article. J Neurosurg 2009;111:265-71. [PubMed]
- Paulus W. GFAP, Ki67 and IDH1: perhaps the golden triad of glioma immunohistochemistry. Acta Neuropathol 2009;118:603-4. [PubMed]
- Yaziji H, Massarani-Wafai R, Gujrati M, et al. Role of p53 immunohistochemistry in differentiating reactive gliosis from malignant astrocytic lesions. Am J Surg Pathol 1996;20:1086-90. [PubMed]
- Myung JK, Cho HJ, Park CK, et al. IDH1 mutation of gliomas with long-term survival analysis. Oncol Rep 2012;28:1639-44. [PubMed]
- Jiao Y, Killela PJ, Reitman ZJ, et al. Frequent ATRX, CIC, FUBP1 and IDH1 mutations refine the classification of malignant gliomas. Oncotarget 2012;3:709-22. [PubMed]
- Sahm F, Reuss D, Koelsche C, et al. Farewell to oligoastrocytoma: in situ molecular genetics favor classification as either oligodendroglioma or astrocytoma. Acta Neuropathol 2014;128:551-9. [PubMed]
- Cairncross G, Jenkins R. Gliomas with 1p/19q codeletion: a.k.a. oligodendroglioma. Cancer J 2008;14:352-7. [PubMed]
- Jiang HH, Ren XH, Zhang Z, et al. A molecular classification system for anaplastic glioma. Zhonghua Wai Ke Za Zhi 2013;51:1104-9. [PubMed]
- Guan X, Vengoechea J, Zheng S, et al. Molecular subtypes of glioblastoma are relevant to lower grade glioma. PLoS One 2014;9:e91216. [PubMed]
- Neill SG, Fisher KE. Section III: Molecular diagnostics in neuro-oncology. Curr Probl Cancer 2014;38:175-9. [PubMed]
- Gupta K, Salunke P. Molecular markers of glioma: an update on recent progress and perspectives. J Cancer Res Clin Oncol 2012;138:1971-81. [PubMed]
- Zhang C, Bao Z, Zhang W, et al. Progress on molecular biomarkers and classification of malignant gliomas. Front Med 2013;7:150-6. [PubMed]
- Le Mercier M, Hastir D, Moles Lopez X, et al. A simplified approach for the molecular classification of glioblastomas. PLoS One 2012;7:e45475. [PubMed]
- Nicolaidis S. Personalized medicine in neurosurgery. Metabolism 2013;62 Suppl 1:S45-8. [PubMed]
- Aldave G, Tejada S, Pay E, et al. Prognostic value of residual fluorescent tissue in glioblastoma patients after gross total resection in 5-aminolevulinic Acid-guided surgery. Neurosurgery 2013;72:915-20; discussion 920-1. [PubMed]
- Hofer S, Rushing E, Preusser M, et al. Molecular biology of high-grade gliomas: what should the clinician know? Chin J Cancer 2014;33:4-7. [PubMed]
- Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006;9:157-73. [PubMed]
- Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010;17:98-110. [PubMed]
- Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010;17:510-22. [PubMed]
- Singh D, Chan JM, Zoppoli P, et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 2012;337:1231-5. [PubMed]
- Ajaz M, Jefferies S, Brazil L, et al. Current and investigational drug strategies for glioblastoma. Clin Oncol (R Coll Radiol) 2014;26:419-30. [PubMed]
- Simon M, Hosen I, Gousias K, et al. TERT promoter mutations: a novel independent prognostic factor in primary glioblastomas. Neuro Oncol 2015;17:45-52. [PubMed]
- Labussière M, Boisselier B, Mokhtari K, et al. Combined analysis of TERT, EGFR, and IDH status defines distinct prognostic glioblastoma classes. Neurology 2014;83:1200-6. [PubMed]
- Malkki H. Neuro-oncology: TERT promoter mutations could indicate poor prognosis in glioblastoma. Nat Rev Neurol 2014;10:546. [PubMed]


