
Correlation of carbonic anhydrase 9 (CA9) with pathological T-stage and prognosis in patients with oral tongue squamous cell carcinoma
Introduction
As a major subtype of head and neck squamous cell carcinoma, oral tongue squamous cell carcinoma (OTSCC) is notable for its remarkable invasiveness, early lymph node metastasis, and poor prognosis with frequent local recurrence (1–3). The quality of life for survivors of OTSCC is often reduced due to difficulties with eating, dysphagia, and speech, as well as paresthesia (4,5). Lymph node and distant metastases are the most reliable adverse prognostic factors in patients with OTSCC (6,7), and despite significant advances in traditional treatment over recent decades, 5-year overall survival (OS) rates and recurrence rates have not markedly improved (8-11). The molecular mechanisms involved in this disease remain incompletely elucidated (12,13), so it is crucial to identify effective biomarkers to improve the diagnosis and prevention of OTSCC.
Gene expression profiling is a powerful method for identifying important mechanistic pathways (14). Microarrays, based on high-throughput platforms, are promising and efficient tools in the search for differentially expressed genes (DEGs) in OTSCC (15). Using integrated and advanced bioinformatic analysis, more reliable and precise results may be obtained from available microarray data, such as those in The Cancer Genome Atlas (TCGA) (16,17), and the Gene Expression Omnibus (GEO) databases (18).
Previous studies on identifying DEGs in OTSCC based on public database and further bioinformatic analysis were rarely reported. In the present study, we performed gene expression profile analysis based on public microarray data from the TCGA to identify key genes. The specific expression of candidate genes were further verified in tumor cells and clinical tumor tissue samples. Our investigation provides insight into the development of OTSCC at the molecular level and identifies candidate prognostic biomarkers for patients with OTSCC. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-7144).
Methods
Microarray data information
Whole-genome mRNA-seq expression data and corresponding clinical information of patients with head and neck SCC were retrieved from the TCGA data portal (https://cancergenome.nih.gov) up to February 14, 2015. The final TCGA mRNA-seq expression data set comprised 20,590 genes from 125 OTSCC tissue samples and 11 adjacent non-tumor tissues. Data were extracted for patients with the tongue as the primary site (mobile tongue), according to the anatomic neoplasm subdivision. Patients with missing survival data were excluded.
An OTSCC mRNA expression dataset (GSE31056) was obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). GSE31056 was generated using the GPL10526 platform (Affymetrix GeneChip Human Genome HG-U133 Plus 2 Array), and included 23 OTSCCs and 73 margin tissues. As these data were obtained from publicly available databases, approval by an ethics committee was not required.
Analysis of gene expression data and clustering
DEGs between tumor tissues and their corresponding adjacent tissues were identified from 11 patients with OTSCC using the empirical Bayes approach in linear models for the limma package in R software. Genes with an expression level of zero (fragments per kilobase of exon model per million reads mapped value) across all patients were omitted. For all P values, the false discovery rate (FDR) was applied for multiple testing correction. Absolute log2 fold-change (FC) ≥1, FDR <0.01, and adjusted P value <0.05 were selected as cut-off criteria. A heat map of DEGs was generated using the heat map package software and showed upregulated and downregulated genes. An online Venn diagram tool (http://bioinfogp.cnb.csic.es/tools/venny/) was used to identify DEGs.
GO and pathway enrichment analysis of DEGs
The Database for Annotation, Visualization, and Integrated Discovery (DAVID, https://david.ncifcrf.gov/), which is a comprehensive set of functional annotation tools, has been used for systematic and integrative analysis of large gene lists (19). Using DAVID, significant GO biological process terms were identified and KEGG pathway enrichment analyses conducted with the identified DEGs as input and a threshold of P<0.05.
PPI network construction
The Search Tool for the Retrieval of Interaction Genes (STRING, http://string-db.org/) database was used for critical assessment and integration of a PPI based on the DEGs; a medium confidence score >0.4 was considered significant (20). Subsequently, a PPI network was reconstructed using Cytoscape software (version 3.6.0, http://www.cytoscape.org/). Next, all significant genes were clustered into several groups to identify important clusters using The Molecular Complex Detection (MCODE) plug-in in Cytoscape (21). The MCODE criteria for selection were as follows: MCODE scores ≥5, degree cut-off =2, node score cut-off =0.2, K−core =2, and max. depth =100.
Survival analysis
OS analysis was performed using 125 OTSCC samples from the TCGA dataset. OS was defined as the time from surgery to death. Data were censored when a patient was alive without recurrence at last follow-up. Patients were classified into high or low expression based on whether gene expression was more (high) or less (low) than the median value. OS analysis of 71 hub genes was performed to evaluate differences between OTSCC samples with varying gene expression. The Cox proportional hazards model was used for multivariate analysis to estimate hazard ratios for individual predictors/covariates; covariates were chosen for inclusion in this analysis if their univariate association with OS had a P value <0.1.
Cell culture and tissue specimens
Cal-27, Tca8113, SCC-9 (human OTSCC cells) and HOK cells (human oral keratinocytes) were purchased from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in a humidified atmosphere of 5% CO2.
Tumor specimens and their corresponding adjacent tissues were collected retrospectively from 20 patients with primary OTSCC treated at the Affiliated Hospital of Qingdao University Medical College between 2016 and 2018. To be eligible for the study, patients had received no prior therapy, and had a primary tumor in the mobile tongue.
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). All patients provided informed consent and the study was approved by the Ethics Committee of the hospital.
Real-time RT-PCR
Total RNA was extracted from Cal-27, Tca8113, SCC-9 and HOK cells using TRIzol reagent (Omega Bio-tek, Norcross, GA, USA). Reverse transcription (RT) was performed using the PrimeScript RT Reagent Kit (TaKaRa, Taejin, Japan). GAPDH was used as an internal control and real-time PCR experiments were repeated three times. The primers for GAPDH were: forward, 5'-ATTCCATGGCACCGTCAAGGCTGA-3' and reverse, 5'-TTCTCCATGGTGGTGAAGACGCCA-3'. The primers for CA9 were: forward, 5'-GTCCAGCTGAATTCCTGCCT-3' and reverse, 5'-CCTTCTGTGCTGCCTTCTCA-3'. All primers were synthesized by Shanghai Sangon Co. (Sangon, Shanghai, China). Real-time PCR was performed using the SYBR Premix Ex Taq II Kit (TaKaRa, Taejin, Japan).
Western blotting
Total proteins were extracted from Cal-27, Tca8113, SCC-9 and HOK cells using prechilled lysis buffer (Sigma-Aldrich, St. Louis, MO, USA). The protein concentration was measured using a BCA Protein Concentration Assay kit (Sigma-Aldrich, St. Louis, MO, USA). Equal amounts of proteins were electrophoresed on 10% SDS-polyacrylamide gels and then transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Boston, MA, USA). After blocking with 5% non-fat milk in Tris-buffered saline for 1 h, membranes were incubated with primary antibody at 4 °C overnight, followed by secondary antibody. Anti-CA9 rabbit polyclonal antibody was purchased from Shanghai Sangon Co. (Sangon, Shanghai, China). The GAPDH protein antibody (Abcam, Cambridge, MA, USA) was selected as an internal reference.
Immunohistochemistry to detect CA9 expression
Tumor and adjacent normal tissues were fixed in 4% formalin and embedded in paraffin. Sections of 4-µm thickness were cut from paraffin blocks, deparaffinized with xylene, and rehydrated in graded alcohol. Antigen retrieval was performed using citrate buffer (pH =6.0). Endogenous peroxidases were blocked with 3% hydrogen peroxide solution. After blocking with 10% normal goat serum to prevent non-specific binding for 30 min, the sections were incubated with anti-CA9 rabbit polyclonal antibody (1:40 dilution, Sangon, Shanghai, China) at 4 °C overnight. Following incubation with labeled Horseradish Peroxidase (HRP) from the Envision kit (Dako, Glostrup, Denmark) for 30 min, color was developed by incubation in 3,3′-diaminobenzidine (DAB) solution. The percentage of positively stained cells was scored as 0 (0%), 1 (1–25%), 2 (26–50%), 3 (51–75%) or 4 (76–100%) by two experienced pathologists. The staining intensity was scored as follows: 0 (no signal), 1 (weak), 2 (moderate) or 3 (strong). The scores for CA9 staining density and intensity were multiplied to obtain a total score, which defined CA9 expression as high (total score ≥3) or low (total score <3).
Statistical analysis
The edge R package (version 3.3.2) was used to screen out DEGs. For OS analysis, the log-rank test was employed using Kaplan-Meier survival curves. Spearman correlation analysis was performed to evaluate the association between gene expression and clinical data. Statistical comparisons were performed using SPSS 12.0 for Windows (SPSS Inc., Chicago, IL, USA), and GraphPad Prism V5.0 software (GraphPadSoftware, La Jolla, CA, USA). Comparisons between groups were carried out using Student’s t-test (two-tailed). Differences were deemed significant if P<0.05.
Results
Identification of DEGs in OTSCC
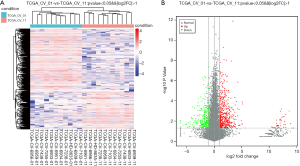
We retrieved expression matrices from 11 pairs of patient tissue samples, comprising cancer and adjacent samples, after data preprocessing and quality assessment using R software. We identified a total of 581 DEGs, including 384 upregulated and 197 downregulated genes. As shown in Figure 1A,B, DEGs were visualized using a volcano plot and a hierarchical-clustering heat map.
Functional enrichment analysis
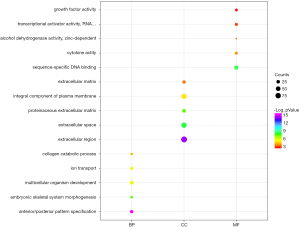
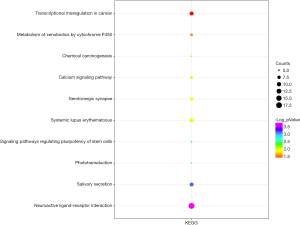
We uploaded all 581 DEGs to the online software, DAVID, to identify overrepresented GO categories and KEGG pathways. The top five GO terms and top 10 KEGG pathways, selected according to P value, are presented in Figures 2 and 3. GO terms in biological processes were enriched for anterior/posterior pattern specification, embryonic skeletal system morphogenesis, and multicellular organism development. Cellular component GO terms were enriched for extracellular region, extracellular space, and proteinaceous extracellular matrix. Molecular function GO terms were most enriched for sequence-specific DNA binding, cytokine activity, alcohol dehydrogenase activity, and zinc-dependent transcriptional activator activity. In addition, the pathway analysis revealed the enrichment of DEGs in many signaling pathways associated with cancer, including neuroactive ligand-receptor interaction, calcium signaling pathway and transcriptional misregulation.
PPI network construction and hub gene identification
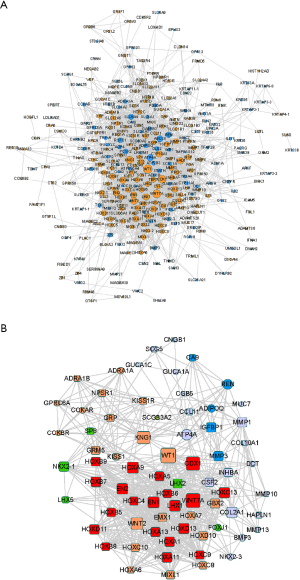
Protein interactions among DEGs were predicted using the online platform, STRING. The PPI network included a total of 301 nodes and 2011 edges and was visualized using Cytoscape (Figure 4A). Subsequently, we used the MCODE plug-in to analyze the whole network and identified five modules, including 71 DEGs (Figure 4B).
Association of DEGs with OS in patients with OTSCC
To evaluate the association of candidate DEG levels with OS in patients with OTSCC, all 71 DEGs were first assessed as independent predictors of OS in the TCGA cohort data. Patients (n=125) were divided into low or high expression groups, according to median gene expression levels (Table 1). The results indicated that only four DEGs were associated with poor OS in patients with OTSCC, namely high expression of CA9, LHX1, and KISS1R, and low expression levels of CCKAR (Figure 5A,B,C,D). We then conducted Kaplan-Meier (log-rank test) univariate survival analysis based on several clinical parameters from the TCGA patient data. The results showed that, besides the key genes described above, pathologic stage (particularly, stage III–IV), pathologic T-stage (stage T3–T4), and smoking history were significant indicators influencing OS of patients with OTSCC (P<0.1). Furthermore, all prognostic factors were analyzed by Cox proportional hazards model multivariate analysis, which showed that only CA9 expression (P=0.029), but not that of the other three genes, and pathological T-stage (P=0.005) were independently associated with OS (Table 2). Remarkably, we noticed that high expression of CA9 was associated with poor pathological T-stage (P=0.033). Overall, the data revealed that CA9 has a vital role in OTSCC and is likely a key molecule affecting prognosis.
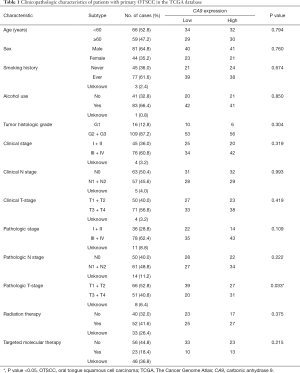
Full table
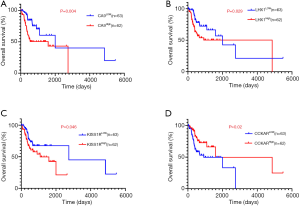
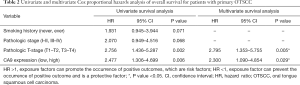
Full table
Identification of a role for CA9 in the biological phenotype of OTSCC
To investigate whether the CA9 molecule may have a specific role in OTSCC, we analyzed a GEO dataset containing 23 OTSCC and 73 margin tissue samples to evaluate the differential expression of CA9 between OTSCC and non-tumor tissues. As shown in Figure 6A, the expression of CA9 in tumor tissue was much higher than in normal tissue, which was consistent with our findings from TCGA analysis.
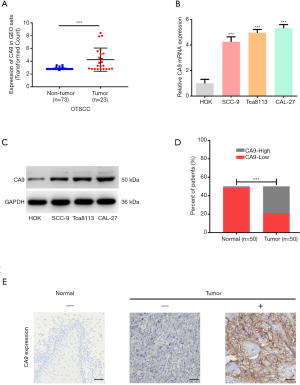
Next, four cell lines, Cal-27, Tca8113, SCC-9 and HOK, were used to compare CA9 expression at both the mRNA and protein levels. As shown in Figure 6B,C, we found that CA9 was overexpressed in cancer cells. Finally, CA9 was stained in clinical tissue samples from 50 patients by immunohistochemistry, and the results also demonstrated that CA9 was highly expressed in tumor cells, relative to normal epithelial cells adjacent to the tumor tissues (Figure 6D,E). Patient characteristics are summarized in Table 3. High CA9 expression was more frequently observed in those with poor pathological T-stage (P=0.038). These results were consistent with TCGA data analysis, and suggested that CA9 exhibits tumor heterogeneity in OTSCC.
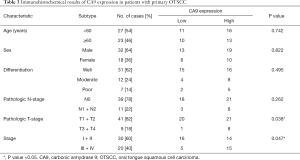
Full table
Discussion
The lethality of OTSCC is mainly because of difficulties in detecting tumors at an early stage and the lack of effective treatments for patients with advanced disease. Biomarkers can be very helpful for the diagnosis, treatment, and prognostic assessment of patients with OTSCC, and bioinformatic analyses play a major role in the investigation and interpretation of genomic and proteomic data (22).In the present study, we identified CA9 as an important tumor specific molecule through bioinformatic screening, supported by molecular biology methods.
Carbonic anhydrase 9 (CA9), a glycoprotein belonging to a family of zinc-containing enzymes, is not highly expressed in most organs or tissues, but is abundantly expressed in numerous cancers and has been investigated as an endogenous marker for tumor hypoxia (23,24). Brockton et al. demonstrated that high (top quartile) stromal CA9 expression was associated with significantly reduced 5-year disease-specific survival, compared with low stromal CA9 expression (25). Other studies support the association of CA9 expression with reduced survival in a number of solid malignancies, including cervical, kidney, and lung cancers (26-29).
Interestingly, whether CA9 affects the prognosis of patients with OTSCC remains controversial. Kim et al. found that high levels of CA9 was an independent prognostic factor in OTSCC (30). Further, Zheng et al. identified the ZEB1-CA9 signaling axis as a biomarker of poor prognosis in tongue cancer (31). On the other hand, Hwa et al. reported that CA9 is not a suitable prognostic marker for OTSCC (32).
Compared with these studies, we were able to draw two reliable conclusions, mainly through the analysis of a OTSCC database, clinical samples, and biological verification. We confirmed that CA9 is a potential prognostic factor in OTSCC, and we also demonstrated that the expression of CA9 was significantly correlated with the pathological T-stage of OTSCC. The T-stage of cancer is when the tumor grows from a single cell to a nodule and then to a tissue. It is also the stage when the tumor moves from being confined to the basement membrane to breaking through it (33). Therefore, we suppose that molecular mechanisms of CA9 may promote OTSCC progression by enhancing tumor invasiveness and clonogenicity. The explicit mechanism by which CA9 is involved in OTSCC requires further exploration.
Besides CA9, there has been recent research on potential biomarkers that are associated with the progression and/or prognosis of OTSCC. VEGF-A and cyclin D1 HIF-1α, SOX2, E-cadherin etc. have been identified as potentially valuable prognostic indicators (34,35), but more specific molecules targeting OTSCC are required, rather than the broad-spectrum molecules expressed in many cancer types.
Conclusions
We identified DEGs and their related pathways and functions by integrated bioinformatic analysis. The results may contribute to elucidating the molecular mechanisms underlying the initiation and development of OTSCC. The CA9 gene has potential to serve as a biomarker and therapeutic target for precise diagnosis and prognostic evaluation of OTSCC.
Acknowledgments
We thank Professor Wei Shang (Department of Stomatology) and the pathologists (Department of Pathology) at the Affiliated Hospital of Qingdao University for their assistance with the immunohistochemical experiments.
Funding: This work was supported by grants from the National Natural Science Foundation of China (grant No. 81703003, ZF) and a China Postdoctoral Science Foundation Funded Project (Project No. 2017M623395 and Project No. 2018T111143).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-7144
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-7144). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of the hospital and all patients provided informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Farquhar DR, Tanner AM, Masood MM, et al. Oral tongue carcinoma among young patients: An analysis of risk factors and survival. Oral Oncol 2018;84:7-11. [Crossref] [PubMed]
- Cunderlikova B, Vasovic V, Randeberg LL, et al. Modification of extracorporeal photopheresis technology with porphyrin precursors. Comparison between 8-methoxypsoralen and hexaminolevulinate in killing human T-cell lymphoma cell lines in vitro. Biochim Biophys Acta 2014;1840:2702-8. [Crossref] [PubMed]
- Kim K, Lee DJ. The updated AJCC/TNM staging system (8th edition) for oral tongue cancer. Transl Cancer Res 2019;8:S164-6.
- Yu X, Li Z. MicroRNA expression and its implications for diagnosis and therapy of tongue squamous cell carcinoma. J Cell Mol Med 2016;20:10-6. [Crossref] [PubMed]
- Bagan J, Sarrion G, Jimenez Y. Oral cancer: clinical features. Oral Oncol 2010;46:414-7. [Crossref] [PubMed]
- Xie N, Wang C, Liu X, et al. Tumor budding correlates with occult cervical lymph node metastasis and poor prognosis in clinical early-stage tongue squamous cell carcinoma. J Oral Pathol Med 2015;44:266-72. [Crossref] [PubMed]
- da Silva SD, Kowalski LP. Perineural invasion in oral cancer: challenges, controversies and clinical impact. Chin Clin Oncol. 2019;8:S5. [Crossref] [PubMed]
- Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin 2012;62:220-41. [Crossref] [PubMed]
- Chen Y, Tian T, Mao MJ, et al. CRBP-1 over-expression is associated with poor prognosis in tongue squamous cell carcinoma. BMC Cancer 2018;18:514. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Li J, Huang H, Sun L, et al. MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin Cancer Res 2009;15:3998-4008. [Crossref] [PubMed]
- Zhang S, Cao R, Li Q, et al. Comprehensive analysis of lncRNA-associated competing endogenous RNA network in tongue squamous cell carcinoma. PeerJ 2019;7:e6397. [Crossref] [PubMed]
- Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol 2008;5:588-99. [Crossref] [PubMed]
- Demokan S, Chuang AY, Chang X, et al. Identification of guanine nucleotide-binding protein gamma-7 as an epigenetically silenced gene in head and neck cancer by gene expression profiling. Int J Oncol 2013;42:1427-36. [Crossref] [PubMed]
- Wang Z, Jensen MA, Zenklusen JC. A Practical Guide to The Cancer Genome Atlas (TCGA). Methods Mol Biol 2016;1418:111-41. [Crossref] [PubMed]
- Lee JS. Exploring cancer genomic data from the cancer genome atlas project. BMB Rep 2016;49:607-11. [Crossref] [PubMed]
- Barrett T, Troup DB, Wilhite SE, et al. NCBI GEO: archive for functional genomics data sets--10 years on. Nucleic Acids Res 2011;39:D1005-10. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [Crossref] [PubMed]
- Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 2017;45:D362-d8. [Crossref] [PubMed]
- Anitha P, Anbarasu A, Ramaiah S. Gene network analysis reveals the association of important functional partners involved in antibiotic resistance: A report on an important pathogenic bacterium Staphylococcus aureus. Gene 2016;575:253-63. [Crossref] [PubMed]
- Wright JT, Hart TC. The genome projects: implications for dental practice and education. J Dent Educ 2002;66:659-71. [Crossref] [PubMed]
- Bussink J, Kaanders JH, van der Kogel AJ. Tumor hypoxia at the micro-regional level: clinical relevance and predictive value of exogenous and endogenous hypoxic cell markers. Radiother Oncol 2003;67:3-15. [Crossref] [PubMed]
- Ivanov S, Liao SY, Ivanova A, et al. Expression of hypoxia-inducible cell-surface transmembrane carbonic anhydrases in human cancer. Am J Pathol 2001;158:905-19. [Crossref] [PubMed]
- Brockton NT, Klimowicz AC, Bose P, et al. High stromal carbonic anhydrase IX expression is associated with nodal metastasis and decreased survival in patients with surgically-treated oral cavity squamous cell carcinoma. Oral Oncol 2012;48:615-22. [Crossref] [PubMed]
- Signoretti S, Regan M, Atkins M. Carbonic anhydrase IX as a predictive biomarker of response to kidney cancer therapy. BJU Int 2008;101 Suppl 4:31-5. [Crossref] [PubMed]
- Kim BW, Cho H, Chung JY, et al. Prognostic assessment of hypoxia and metabolic markers in cervical cancer using automated digital image analysis of immunohistochemistry. J Transl Med 2013;11:185. [Crossref] [PubMed]
- Giatromanolaki A, Koukourakis MI, Sivridis E, et al. Expression of hypoxia-inducible carbonic anhydrase-9 relates to angiogenic pathways and independently to poor outcome in non-small cell lung cancer. Cancer Res 2001;61:7992-8. [PubMed]
- Koukourakis MI, Giatromanolaki A, Sivridis E, et al. Hypoxia-regulated carbonic anhydrase-9 (CA9) relates to poor vascularization and resistance of squamous cell head and neck cancer to chemoradiotherapy. Clin Cancer Res 2001;7:3399-403. [PubMed]
- Kim SJ, Shin HJ, Jung KY, et al. Prognostic value of carbonic anhydrase IX and Ki-67 expression in squamous cell carcinoma of the tongue. Jpn J Clin Oncol 2007;37:812-9. [Crossref] [PubMed]
- Zheng G, Peng C, Jia X, et al. ZEB1 transcriptionally regulated carbonic anhydrase 9 mediates the chemoresistance of tongue cancer via maintaining intracellular pH. Mol Cancer 2015;14:84. [Crossref] [PubMed]
- Hwa JS, Kwon OJ, Park JJ, et al. The prognostic value of immunohistochemical markers for oral tongue squamous cell carcinoma. Eur Arch Otorhinolaryngol 2015;272:2953-9. [Crossref] [PubMed]
- Almangush A, Mäkitie AA, Triantafyllou A, et al. Staging and grading of oral squamous cell carcinoma: An update. Oral Oncol 2020;107:104799. [Crossref] [PubMed]
- Almangush A, Heikkinen I, Mäkitie AA, et al. Prognostic biomarkers for oral tongue squamous cell carcinoma: a systematic review and meta-analysis. Br J Cancer 2017;117:856-66. [Crossref] [PubMed]
- Hussein AA, Forouzanfar T, Bloemena E, et al. A review of the most promising biomarkers for early diagnosis and prognosis prediction of tongue squamous cell carcinoma. Br J Cancer 2018;119:724-36. [Crossref] [PubMed]
(English Language Editor: K. Brown)


