
KIF23 enhances cell proliferation in pancreatic ductal adenocarcinoma and is a potent therapeutic target
Introduction
In recent years, significant progress has been made in terms of cancer research and treatment. However, there has not been any remarkable advance in survival rates for pancreatic cancer (1,2). The morbidity and mortality of pancreatic ductal adenocarcinoma (PDAC) have risen significantly in recent years: with 56,770 new cases (male 29,940 and female 26,830 cases) of pancreatic cancer and 45,750 deaths (male 23,800 and female 21,950 cases) expected in 2019 (3), 55,400 new cases (male 29,200 and female 26,240 cases) and 44,330 deaths (male 23,020 and female 21,310 cases) reported in 2018 (4), and 53,670 new cases (male 27,970 and female 25,700 cases) and 43,090 (male 22,300 and female 20,790 cases) deaths in 2017 (5). The five-year survival rate for pancreatic cancer is very low, and it is considered one of the worst malignant tumors due to its high surgical mortality rate and low cure rate (6). In China, the mortality rate from pancreatic cancer is almost equal to the incidence rate because of the poor prognosis (7). Although there are treatment options for pancreatic cancer, such as chemotherapy and radiotherapy, surgical resection is still the main treatment of choice. However, most patients with pancreatic cancer are diagnosed at an advanced stage and surgery is feasible in less than 20% of cases (8). Recent studies have found that the occurrence of pancreatic cancer is largely affected by gender, aging, smoking, and diabetes (9). However, the pathogenesis of pancreatic cancer is complex and our understanding remains limited. Therefore, more in-depth studies on the mechanism of pancreatic cancer may help us to identify novel therapeutic targets (10).
Kinesin family member 23 (KIF23), a human homolog of mouse KIF23, is one of the kinesin motor proteins involved in the regulation of cytokinesis (11,12). Based on cDNA microarray or by quantitative reverse transcription-polymerase chain reaction (PCR), KIF23 overexpression has been detected in lung cancer, breast cancer, gliomas, paclitaxel-resistant gastric cancer, hepatocellular carcinoma, and pancreatic cancer (13,14), and has been associated with poor prognosis for several cancers (15).
In our research, we discovered that high expression of KIF23 was associated with poor prognosis by immunohistochemical analysis of tumor sections and clinical data from patients with PDAC. We also found that knockdown of KIF23 decreased proliferation in PANC-1 and BxPC-3 PDAC cells, and suppression of KIF23 in PANC-1 cells inhibited tumor growth in nude mice. We present the following article in accordance with the ARRIVE reporting checklist (available at http://dx.doi.org/10.21037/atm-20-1970).
Methods
Bioinformatic analysis
We used GEPIA (http://gepia.cancer-pku.cn/) (16) to collate and analyze The Cancer Genome Atlas (TCGA) data with a threshold of P<0.05 and LogFC >1 or <−1 for the differential genes, and we divided the patients into two groups using the median value as the basis for Kaplan-Meier survival analysis. The 95% confidence interval is marked with a dotted line. In addition, the correlation between KIF23 and CDCA8 was analyzed by GEPIA.
Sample collection
Based on the institutional review board's approval, all of the 82 adult PDAC patients with PDAC included in this study were from Tianjin Medical University Cancer Institute and Hospital from 2009 to 2017. These cases were included on the basis of a clear pathological diagnosis. The informed consent of each patient was obtained. All procedures performed in research involving human participants complied with the ethical standards of the institution and/or the National Research Council and with the 1964 Helsinki Declaration and its subsequent amendments or similar ethical standards.
Immunohistochemistry
The expression of KIF23 was detected in paraffin-embedded tumor tissue by a rabbit-anti-KIF23 antibody (ab235955, 1:200 dilution; Abcam, USA), following the procedure specified in the immunohistochemistry kit (ZSGB-BIO, pv6000, China). Briefly, we fixed tumor tissues and matched normal adjacent tissues in 10% neutral-buffered formalin and embedded them in paraffin blocks by the pathology department of our hospital. Then, the paraffin sections (4 µm) were placed into a microwave oven for 15 minutes to repair the antigens and were then cooled at room temperature. Endogenous peroxidase was blocked, and sections were incubated with an anti-KIF23 antibody overnight at 4 °C, followed by a goat anti-rabbit secondary antibody at 37 °C for 1 hour. Sections were stained with 3,3-diaminobenzidin (DAB) at room temperature for 10 minutes and images were then recorded by microscopy (Olympus BX43). All samples were evaluated by two independent pathologists.
To analyze the results, KIF23 nuclear staining was scored using the 4-point scale (0, no staining; 1+, light staining at high magnification; 2+, intermediate staining; and 3+, dark staining of the linear membrane at low magnification). According to the distribution of scores of KIF23 staining intensity, samples were divided into high expression (2+ to 3+) and low expression (0 to 1+) groups.
Antibodies
The antibodies used in this study were: rabbit anti-KIF23 antibody (Abcam, ab235955; 1:200 dilution for immunohistochemistry and 1:1000 dilution for western blotting), rabbit anti-β-actin antibody (1:1000 dilution for western blotting; ab8227, Abcam plc., Cambridge, UK), rabbit anti-Ki67 antibody (1:200 dilution for immunohistochemistry and 1:1000 dilution for western blotting; ab16667, Abcam), rabbit anti-proliferating cell nuclear antigen (PCNA) antibody (1:500 dilution for western blotting; ab18197, Abcam), and rabbit anti-Borealin/CDCA8 (ab70910, Abcam; 1:50 dilution for immunohistochemistry).
RNA isolation and qPCR
Total RNA was isolated from cancer cells using TRIzol reagent (Invitrogen, 15596026, USA) and complementary DNA (cDNA) synthesis was performed using the PrimeScript RT Reagent Kit (Clontech, RR055B, USA). The following primers were used in the reactions: KIF23 (forward primer: 5'-CAGATTTCCAACGGCCAGCA-3' and reverse primer: 5'-TGGCTTTTTGCGCTTGG-3'), and GAPDH (forward primer: 5'-CGACCACTTTGTCAAGCTCA-3' and reverse primer: 5'-GGTTGAGCACAGGGTACTTTATT-3ʹ).
Western blot analysis
Total protein was collected from PANC-1 and BxPC-3 cells using RIPA and RMSF reagents (100:1) on ice and was then quantified by a BCA assay. Proteins were separated on 10% PAGE gels, then transferred to PVDF membrane (Millipore, IPSN07852, USA). The membrane was washed three times using PBST and blocked with 10% nonfat milk for 1 hour, then incubated with a primary antibody at 4 °C overnight. After a further three washes, PVDF was incubated with a secondary antibody for 1 hour at room temperature and then washed again. Finally, the ECL substrate reagent kit (GE Healthcare, RPN 2109, USA) was used to visualize the protein bands on the imaging capture system (Alpha Innovation).
Cell culture
The PDAC cells of PANC-1 and BxPC-3 were cultured in 1640 medium (Gibco, 61870044, USA) with 10% fetal bovine serum (Gibco, 10099-141, USA) at 37 °C in a 5% CO2 incubator.
The shRNA and stable clones
Lenti-KD RNAi (GICL3071096830, Genechem, China; AAACGAACCTTAAAGACCCAGTT) against KIF23 was transfected into PANC-1 and BxPC-3 PDAC cells using Lipofectamine e3000 reagent (Invitrogen, L30000315, CA, USA). The KIF23 shRNA control sequence plasmids were transfected as a negative control. Six-well plates were seeded with 1×105 cells per well based on the manufacturer's protocol. Wells were divided into three groups: the KIF23 depletion group, which was transfected with shRNA targeting KIF23; a negative control (NC) group, which was transfected with the control sequence; and a Mock group, which was treated without any reagent (data not shown). Silencing efficiency was measured by both quantitative PCR and immunoblotting 48 hours after transfection. The depleted cells were used to evaluate the correlation between KIF23 expression and cellular processes. Cells transfected with shRNA targeting KIF23 were maintained in medium containing 2 µg/mL puromycin for 2 weeks to select stable clones. Then, stable KIF23-depleted cell lines were screened through lentivirus infection and used for the both in vitro and in vivo assays.
Colony formation assays
PANC-1 and BxPC-3 cells were seeded into six-well plates (500 cells/well) and incubated at 37 °C in 5% CO2 for 2 weeks. Cells were fixed with 100% methanol and stained with 0.1% crystal violet. Images were collected during microscopy, and quantitation was performed using Image-Pro Plus 5.0 software.
MTT assay
An MTT assay was used to evaluate cell proliferation and survival. Cells were resuspended in DMEM medium and 200 µL of cell suspension was seeded into 96-well plates at 2,000 cells/well and incubated at 37 °C in 5% CO2. After three days, the medium was removed and 20 µL of MTT solution was added into each well and further incubated at 37 °C for 4 hours. Then, dimethyl sulfoxide (DMSO) was added (150 µL/well) to solubilize the formazan crystals, and the plate was incubated at room temperature for 10 minutes on a shaking table. The absorbance was then measured at 570 nm by spectrophotometry.
Mouse xenograft model
Nude BALB/c mice (female, 6–8 weeks, 18–22 g, four mice in each group) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). PANC-1 PDAC cells (5×106, 100 µL) were injected subcutaneously into the armpit of the nude mice. The size of tumors was measured every 3 days, and the tumor volume was calculated using the formula V = 0.5 × L × W2, where V is the volume, L is the length, and W is the width. After 29 days, the tumors were harvested, weighed, and photographed. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. Animal research was performed according to guidelines approved by the Tianjin Medical University Cancer Institute and the Hospital Animal Ethics Committee (approval No.: IRM-DWLL-2019113).
Semi-quantitative analysis of the immunohistochemical data from in vivo tumors
The expression of KIF23 or Ki67 was detected in paraffin-embedded tumors using a rabbit-anti-KIF23 or rabbit-anti-Ki67 antibody, following the procedure detailed in the immunohistochemistry kit (ZSGB-BIO, pv6000, China). Briefly, the pathology department of our hospital excised the tumor tissues in vivo and fixed them in 10% neutral-buffered formalin and then embedded them in paraffin blocks. Then, the paraffin-embedded sections (4 µm) were placed into a microwave oven for 15 minutes to repair the antigens and were then cooled at room temperature. Endogenous peroxidase was blocked and sections were incubated with an anti-KIF23 (or Ki67) antibody overnight at 4 °C and then a goat anti-rabbit antibody at 37 °C for 1 hour. Sections were stained with DAB at room temperature for 10 minutes. Images were obtained under microscopy (Olympus BX43). Then, positive cells were counted in five randomly selected fields under a light microscope at a 200× magnification, and the data were expressed as the mean value ± standard deviation (SD).
Statistical methods
Statistical analyses were conducted using SPSS edition 22.0. The relationship between KIF23 expression and clinical features was assessed using a chi-square test. The Kaplan-Meier method were employed to analyze disease-free survival and overall survival in pancreatic cancer patients. A t-test was performed for comparison of the data between the groups. Data were expressed as the mean value ± SD. Two-sided P values of <0.05 were considered significant.
Results
High expression of KIF23 was related to poor prognosis
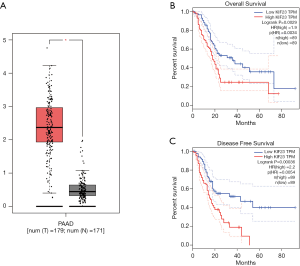
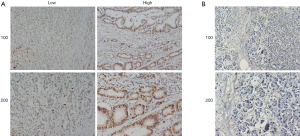
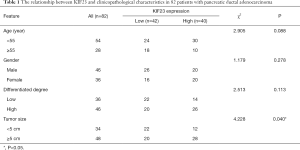
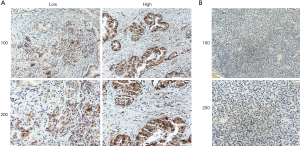
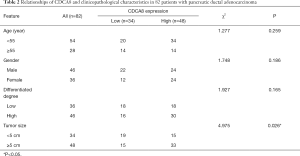
A bioinformatic method was used to analyze the expression of KIF23 at the mRNA level in pancreatic tumor tissues and normal pancreatic tissues. As shown in Figure 1, KIF23 mRNA expression was higher in pancreatic cancer tissues than in normal tissues (Figure 1A, P<0.05). As shown in Figure 1B and 1C, the median KIF23 mRNA expression level was used as the cut-off point to divide all pancreatic cancer patients into two groups: high expression (n=89, from the TCGA dataset) and low expression (n=89, from the TCGA dataset). Patients in the high expression group showed worse overall survival and relapse free survival rates. We also examined the expression of KIF23 in peripheral blood and tumors. We found that KIF23 has guiding significance for the prognosis of pancreatic cancer, and KIF23 was also expressed in tumors (Figure S1). These data suggested that KIF23 might be a poor prognostic factor in pancreatic cancer patients. To verify these results, we conducted immunohistochemical studies to investigate the expression of KIF23 protein in PDAC patients from our institute. As shown in Figure 2A, KIF23 positive staining was localized in the nucleus. Weak or no reactivity was observed in the low expression samples, whereas strong staining was observed in the high expression samples. Compared to Figure 2A, the KIF23 negative staining was observed in normal tissue adjacent to the cancer tissue in Figure 2B. We also analyzed the connection between KIF23 expression and the clinical features of pancreatic cancer patients. As shown in Table 1, high expression of KIF23 was related to tumor size (P=0.04). However, high expression of KIF23 was not related to age (P=0.088), gender (P=0.278), or the degree of differentiation (P=0.113).
Full table
KIF23 knockdown in PANC-1 and BxPC-3 PDAC cells by short hairpin RNA (shRNA)
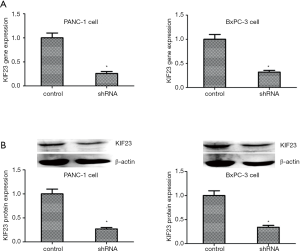
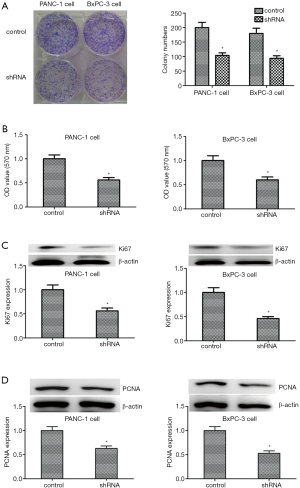
First, to observe the function of KIF23 in PDAC, PANC-1 and BxPC-3 cells were transfected with specific shRNA to knockdown KIF23. Total RNA was isolated from the cells and the expression of the gene encoding KIF23 was analyzed by PCR. As shown in Figure 3A, when PANC-1 and BxPC-3 cells were transfected with shRNA, the low expression of KIF23 was detected by PCR. The control cells were transfected with empty plasmid. Second, we used western blotting to analyze the KIF23 protein expression levels. The results were consistent with those for the RNA levels (Figure 3B). The PANC-1 and BxPC-3 cells transfected with shRNA were selected with puromycin until the cells formed stable clones.
Knockdown of KIF23 inhibited the proliferation of PANC-1 and BxPC-3 cells
To study the influence of KIF23 on PDAC cells, a colony formation assay was conducted and KIF23 was knocked down by shRNA in PANC-1 and BxPC-3 PDAC cells. Compared with the control cells, the proliferation of PDAC cells was inhibited following knockdown of KIF23 (Figure 4A). Moreover, the results of the MTT assay were similar to those obtained with the colony formation assay (Figure 4B). Ki67 and PCNA are two markers of cell proliferation. When transfected with KIF23 shRNA, the expression levels of Ki67 and PCNA in PANC-1 cells and in BxPc-3 cells were lower (Figure 4C,D). These data indicated that KIF23 affects the proliferation of PANC-1 and BxPc-3 cells.
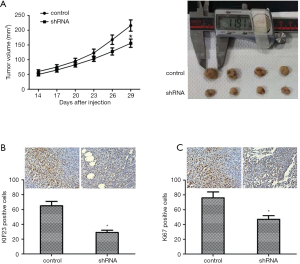
Knockdown of KIF23 inhibited tumor growth in vivo
To study the effect of KIF23 in vivo, PANC-1 cells transfected with KIF23 shRNA were injected into nude mice subcutaneously. Tumors were measured every 3 days and excised after 29 days. As shown in Figure 5A, tumor growth in the shKIF23 group was significantly inhibited. Then, we used immunohistochemistry to detect the expression of KIF23 in tumors. The expression level of KIF23 protein in the control group was obviously higher than that in tumors (Figure 5B). These data showed that KIF23 was successfully knocked down in the shKIF23 group. The expression level of Ki67 in the control group was also higher than in tumors (Figure 5C). These results revealed that KIF23 is a positive regulator of PDAC progression.
KIF23 promotes pancreatic cancer proliferation by affecting CDCA8
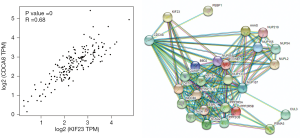
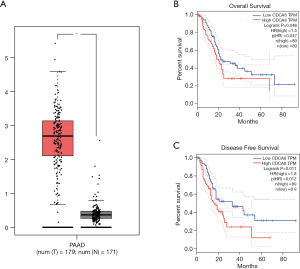
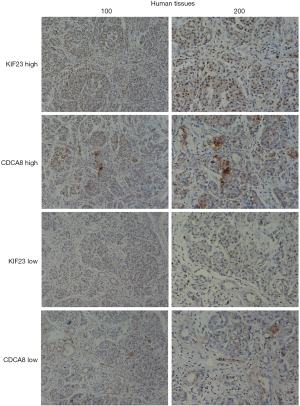
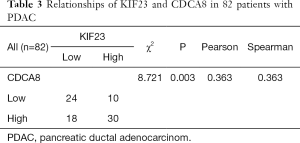
Based on our finding that KIF23 can regulate cell proliferation of pancreatic cancer both in vitro and in vivo, we next investigated the potential mechanism responsible for the involvement of KIF23 in tumor growth. A bioinformatic method was used to analyze the relationship between KIF23 and CDCA8 (http://gepia.cancer-pku.cn/detail.php?gene=KIF23), and a strong positive correlation was detected (R=0.68, P=0.000<0.05, Figure 6A). Then, using GeneCards software, positive gene interactions between KIF23 and CDCA8 were revealed (https://www.genecards.org/cgi-bin/carddisp.pl?gene=KIF23, Figure 6B). After confirming the expression of CDCA8 (https://www.proteinatlas.org/ENSG00000134690-CDCA8/pathology), we concluded that the expression of CDCA8 in the peripheral blood during pancreatic cancer has guiding significance for prognosis (Figure S1), and that this protein is expressed in tumors. At the same time, we found that KIF23 has similar situation in tumors (Figure S2). After checking GEPIA (http://gepia.cancer-pku.cn/), the mRNA expression of CDCA8 was confirmed to be significantly higher in tumor tissue than in normal pancreatic tissue. Taking the median as the cutoff value, the patients were divided into high and low expression groups. The results showed that the OS (overall survival) and RFS (relapse free survival) rates of the high expression group were poorer (P<0.05, Figure 7). Then, we performed immunohistochemistry to analyze the protein expression of CDCA8, and strong staining was observed for the high expression group compared with weak staining for the low expression group (Figure 8). Moreover, we analyzed the relationship between the expression of CDCA8 and the clinicopathological characteristics of pancreatic cancer, and found that high expression was related to tumor size (P=0.026<0.05), but aging (P=0.259), gender (P=0.186) and differentiation (P=0.165) showed no correlation (Table 2). These results were similar to those of KIF23. Moreover, immunohistochemical analysis of the tumor tissues revealed that low CDCA8 expression correlated with low expression of KIF23, and vice versa (Figure 9). Correlation analysis on our data confirmed the significant positive correlation between the expression of CDCA8 and KIF23 (P=0.003<0.05) (Table 3). According to these results, we propose that KIF23 promotes pancreatic cancer by affecting the CDCA8 signaling pathway; however, further verification of this mechanism is necessary.
Full table
Full table
Discussion
Pancreatic cancer, known as “the king of cancer”, is hard to diagnose and treat and is therefore associated with among the worst prognoses of all malignant tumors (16-22). In recent years, the morbidity and mortality rates for pancreatic cancer have markedly increased, and the 5-year survival rate is just less than 1% (18-27). Currently, the treatment methods for pancreatic cancer mainly include chemotherapy, radiotherapy, surgical treatment, and targeted treatment, but most have little effect (21-25). Although current pancreatic cancer therapeutic targets have achieved certain therapeutic effects, novel effective therapeutic targets are urgently needed and thereby, a deeper understanding of the pathogenesis of pancreatic cancer is required (21-35).
To the best of our knowledge, this is the first report of KIF23 expression in pancreatic cancer. We report that the expression of KIF23 differs among PDAC patients, and high expression of KIF23 is related to a poor prognosis. Our results suggest that KIF23 may be a new prognosis factor in PDAC.
Nislow first identified a human KIF23 homolog in 1992 (13), which was a plus-end-directed motor enzyme that translocates antiparallel microtubules in vitro. Following this, other reports confirmed that KIF23 as a key regulator of cytokinesis (12,36-40). By screening a HeLa cell cDNA library, KIF23 (also known as mitotic kinesin-like protein-1, MKLP1) was separated and confirmed as a plus-end-directed motor enzyme that translocates antiparallel microtubules in vitro (41). This enzyme contains the kinesin superfamily motor domain at its N-terminus, and has been found to be localized in both the cytoplasm and nucleus (42). The kinesin motor protein KIF23 was reported to be a underlying marker for several tumors (13). KIF23 is essential for the completion of cytokinesis in midbody formation and for recruitment to the spindle midzone/midbody by a chromosomal passenger protein (40). Liu and Erikson reported that a KIF23 mutant lacking nuclear localization signals, led to cell cycle arrest, further demonstrating that KIF23 is essential for cytokinesis (43). In addition, KIF23 acts as the motor enzyme for microtubules, which are known to play an important role in cellular shape and processes such as motility, mitosis, intracellular vesicle transport, organization, and positioning of membranous organelles (44-47). KIF23 overexpression is a feature of some tumors, such as glioma (48), breast (49), and paclitaxel-resistant gastric cancer (50). Our study also showed that KIF23 was overexpressed in PDAC patients. Importantly, high levels of KIF23 mRNA correlated with poor survival in patients with glioma and ER-positive breast cancer (48,50). In our study, KIF23 was demonstrated to play a vital role in PDAC cell proliferation. Our study analyzed the function of KIF23 in PDAC. PANC-1 and BxPC-3 cells were transfected with specific shRNA to KIF23 and the resulting phenotypes were observed. The results of an MTT assay and the expression level of PCNA revealed that knockdown of KIF23 with shRNA can inhibit the proliferation of PDAC cells, which was consistent with other reports in glioma (48), breast (49), and paclitaxel-resistant gastric cancer (50). Compared with the cells transfected with control shRNA, the stable clone of KIF23-knockdown cells grew slower in vivo. Based on these data, KIF23 may be an oncogene in PDAC.
There are some limitations in our study. It is retrospective and includes only a small number of patient samples from a single center; however, few previous studies have investigated the detailed mechanism by which KIF23 regulates tumor growth in PDAC. We reported a positive correlation between KIF23 and CDCA8 based on TCGA data (GEPIA and Genecards) and immunohistochemistry (our clinical data) further verification may be required in future studies. Furthermore, our conclusion that KIF23 may promote pancreatic cancer proliferation by affecting the expression of CDCA8 in the pathway, may also require further verification in the future.
In conclusion, our study indicated that KIF23 might play a critical role in the progression of PDAC and that it operates through enhanced cell proliferation, which was consistent with the findings of other previous studies. Further, a positive correlation between KIF23 and CDCA8 was demonstrated, and KIF23 might promote pancreatic cancer proliferation by affecting CDCA8 expression. These findings may prompt further studies of KIF23 as a therapeutic target for the treatment of PDAC. However, these findings are preliminary and further studies are needed to confirm these results.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (Grant no: 81401957). At the same time this work was supported by Tianjin Key Laboratory of Radiation Medicine and Molecular Nuclear Medicine, Institute of Radiation Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College.
Footnote
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-1970
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-1970
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-1970
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-1970). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in research involving human participants complied with the ethical standards of the institution and/or the National Research Council and with the 1964 Helsinki Declaration and its subsequent amendments or similar ethical standards. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. Animal research was performed according to guidelines approved by the Tianjin Medical University Cancer Institute and the Hospital Animal Ethics Committee (approval No.: IRM-DWLL-2019113). Our protocol was approved by the committee and all operations were performed under sodium pentobarbital anesthesia, with every effort made to minimize pain.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Khan MA, Azim S, Zubair H, et al. Molecular drivers of pancreatic cancer pathogenesis: looking inward to move forward. Int J Mol Sci 2017;18:779. [Crossref] [PubMed]
- Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin 2017;67:177-93. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics. 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics. 2018. CA Cancer J Clin 2018;68:7-30. [Crossref] [PubMed]
- Siegel RL, Miller KD, Jemal A. Cancer statistics. 2017. CA Cancer J Clin 2017;67:7-30. [Crossref] [PubMed]
- Neoptolemos JP, Stocken DD, Bassi C, et al. Adjuvant chemotherapy with fluorouracil plus folinic acid vs gemcitabine following pancreatic cancer resection: a randomized controlled trial. JAMA 2010;304:1073-81. [Crossref] [PubMed]
- Chen W, Zheng R, Zuo T, et al. National cancer incidence and mortality in China, 2012. Chin J Cancer Res 2016;28:1-11. [PubMed]
- Sharma C, Eltawil KM, Renfrew PD, et al. Advances in diagnosis, treatment and palliation of pancreatic carcinoma: 1990-2010. World J Gastroenterol 2011;17:867-97. [Crossref] [PubMed]
- Vincent A, Herman J, Schulick R, et al. Pancreatic cancer. Lancet 2011;378:607-20. [Crossref] [PubMed]
- Sun X, Kong L, Li B, et al. Peroxiredoxin 1 silencing inhibited the growth and promoted apoptosis of pancreatic cancer cells via targeting FOXO3 gene. Cancer Manag Res 2018;10:5019-26. [Crossref] [PubMed]
- Glotzer M. The molecular requirements for cytokinesis. Science 2005;307:1735-9. [Crossref] [PubMed]
- Neef R, Klein UR, Kopajtich R, et al. Cooperation between mitotic kinesins controls the late stages of cytokinesis. Curr Biol 2006;16:301-7. [Crossref] [PubMed]
- Nislow C, Lombillo VA, Kuriyama R, et al. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature 1992;359:543-7. [Crossref] [PubMed]
- Zhou Z, Cheng Y, Jiang Y, Liu S, Zhang M, Liu J, et al. Ten hub genes associated with progression and prognosis of pancreatic carcinoma identified by co-expression analysis. Int J Biol Sci 2018;14:124-36. [Crossref] [PubMed]
- Vikberg AL, Vooder T, Lokk K, Annilo T, et al. Mutation analysis and copy number alterations of KIF23 in non-small-cell lung cancer exhibiting KIF23 over-expression. Onco Targets Ther 2017;10:4969-79. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Sun Y, Peng W, He W, et al. Transgelin-2 is a novel target of KRAS-ERK signaling involved in the development of pancreatic cancer. J Exp Clin Cancer Res 2018;37:166. [Crossref] [PubMed]
- Tohme S, Kaltenmeier C, Bou-Samra P, et al. race and health disparities in patient refusal of surgery for early-stage pancreatic cancer: an NCDB cohort study. Ann Surg Oncol 2018;25:3427-35. [Crossref] [PubMed]
- Tannapfel A. Pancreatic cancer. Molecular and surgical pathology Pathologe. 2010;31:225-8. [PubMed]
- Kong B, Michalski CW, Erkan M, Friess H, Kleeff J. From tissue turnover to the cell of origin for pancreatic cancer. Nat Rev Gastroenterol Hepatol 2011;8:467-72. [Crossref] [PubMed]
- Borazanci E, Dang CV, Robey RW, et al. Pancreatic cancer: “a riddle wrapped in a mystery inside an enigma Clin Cancer Res 2017;23:1629-37. [Crossref] [PubMed]
- Chin V, Nagrial A, Sjoquist K, et al. Chemotherapy and radiotherapy for advanced pancreatic cancer. Cochrane Database Syst Rev 2018;3:CD011044. [Crossref] [PubMed]
- Chen KT, Devarajan K, Milestone BN, et al. Neoadjuvant chemoradiation and duration of chemotherapy before surgical resection for pancreatic cancer: does time interval between radiotherapy and surgery matter? Ann Surg Oncol 2014;21:662-9. [Crossref] [PubMed]
- Yang SH, Kuo TC, Wu H, et al. Perspectives on the combination of radiotherapy and targeted therapy with DNA repair inhibitors in the treatment of pancreatic cancer. World J Gastroenterol 2016;22:7275-88. [Crossref] [PubMed]
- Lan C, Li X, Wang X, et al. A new combined criterion to better predict malignant lesions in patients with pancreatic cystic neoplasms. Cancer Biol Med 2018;15:70-8. [Crossref] [PubMed]
- Zeng S, Pöttler M, Lan B, et al. Chemoresistance in pancreatic cancer. Int J Mol Sci 2019;20:E4504. [Crossref] [PubMed]
- Ayhan A, Kuhn E, Wu RC, et al. CCNE1 copy-number gain and overexpression identify ovarian clear cell carcinoma with a poor prognosis. Mod Pathol 2017;30:297-303. [Crossref] [PubMed]
- Huang HN, Huang WC, Lin CH, et al. Chromosome 20q13.2 ZNF217 locus amplification correlates with decreased E-cadherin expression in ovarian clear cell carcinoma with PI3K-Akt pathway alterations. Hum Pathol 2014;45:2318-25. [Crossref] [PubMed]
- Forbang RT, Srinivasan S, Ng SP, et al. Setup management for stereotactic body radiation therapy of patients with pancreatic cancer treated via the breath-hold technique. Pract Radiat Oncol 2020;10:e280-e289. [Crossref] [PubMed]
- Müller D, Shin S, Goullet de Rugy T, et al. eIF4A inhibition circumvents uncontrolled DNA replication mediated by 4E-BP1 loss in pancreatic cancer. JCI Insight 2019;4:121951. [Crossref] [PubMed]
- Carr RM, Fernandez-Zapico ME. Toward personalized TGFβ inhibition for pancreatic cancer. EMBO Mol Med 2019;11:e11414. [Crossref] [PubMed]
- Rossi ML, Rehman AA, Gondi CS. Therapeutic options for the management of pancreatic cancer. World J Gastroenterol 2014;20:11142-59. [Crossref] [PubMed]
- McGuigan A, Kelly P, Turkington RC, et al. Pancreatic cancer: a review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol 2018;24:4846-61. [Crossref] [PubMed]
- Jahan R, Ganguly K, Smith LM, et al. Trefoil factor(s) and CA19.9: a promising panel for early detection of pancreatic cancer. EBioMedicine 2019;42:375-85. [Crossref] [PubMed]
- Liu J, Gao J, Du Y, et al. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int J Cancer 2012;131:683-91. [Crossref] [PubMed]
- Chen MC, Zhou Y, Detrich HW 3rd. Zebrafish mitotic kinesin-like protein 1 (Mklp1) functions in embryonic cytokinesis. Physiol Genomics 2002;8:51-66. [Crossref] [PubMed]
- Kuriyama R, Gustus C, Terada Y, et al. CHO1, a mammalian kinesin-like protein, interacts with F-actin and is involved in the terminutes al phase of cytokinesis. J Cell Biol 2002;156:783-90. [Crossref] [PubMed]
- Li WM, Webb SE, Chan CM, et al. Multiple roles of the furrow deepening Ca2+ transient during cytokinesis in zebrafish embryos. Dev Biol 2008;316:228-48. [Crossref] [PubMed]
- Seguin L, Liot C, Mzali R, et al. CUX1 and E2F1 regulate coordinated expression of the mitotic complex genes Ect2, MgcRacGAP, and MKLP1 in S phase. Mol Cell Biol 2009;29:570-81. [Crossref] [PubMed]
- Zhu C, Bossy-Wetzel E, Jiang W. Recruitment of MKLP1 to the spindle midzone/midbody by INCENP is essential for midbody formation and completion of cytokinesis in human cells. Biochem J 2005;389:373-81. [Crossref] [PubMed]
- Kato T, Wada H, Patel P, et al. Overexpression of KIF23 predicts clinical outcome in primary lung cancer patients. Lung Cancer 2016;92:53-61. [Crossref] [PubMed]
- Schmidt EE, Pelz O, Buhlmann S, et al. GenomeRNAi: a database for cell-based and in vivo RNAi phenotypes, 2013 update. Nucleic Acids Res 2013;41:D1021-6. [Crossref] [PubMed]
- Liu X, Erikson RL. The nuclear localization signal of mitotic kinesin-like protein Mklp-1: effect on Mklp-1 function during cytokinesis. Biochem Biophys Res Commun 2007;353:960-4. [Crossref] [PubMed]
- Calligaris D, Verdier-Pinard P, Devred F, et al. Microtubule targeting agents: from biophysics to proteomics. Cell Mol Life Sci 2010;67:1089-104. [Crossref] [PubMed]
- Kline-Smith SL, Walczak CE. Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol Cell 2004;15:317-27. [Crossref] [PubMed]
- Vasiliev JM, Gelfand IM, Domnina LV, et al. Effect of colcemid on the locomotory behaviour of fibroblasts. J Embryol Exp Morphol 1970;24:625-40. [PubMed]
- Wittmann T, Hyman A, Desai A. The spindle: a dynamic assembly of microtubules and motors. Nat Cell Biol 2001;3:E28-34. [Crossref] [PubMed]
- Takahashi S, Fusaki N, Ohta S, et al. Downregulation of KIF23 suppresses glioma proliferation. J Neurooncol. 2012;106:519-29. [Crossref] [PubMed]
- Zou JX, Duan Z, Wang J, et al. Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival, and tamoxifen resistance. Mol Cancer Res 2014;12:539-49. [Crossref] [PubMed]
- Murakami H, Ito S, Tanaka H, et al. Establishment of new intraperitoneal paclitaxel-resistant gastric cancer cell lines and comprehensive gene expression analysis. Anticancer Res 2013;33:4299-307. [PubMed]


