
Comprehensive analysis of circular RNA in oral leukoplakia: upregulated circHLA-C as a potential biomarker for diagnosis and prognosis
Introduction
Oral leukoplakia (OLK) is predominantly a whitish patch or plaque that cannot be characterized clinically or pathologically as any other disease (1). Epidemiology shows that the incidence of malignant transformation is as high as 3–18%, and can be as high as 15–40% for heterogeneous OLK (2). Due to its complex etiology, the pathogenesis of OLK is still unclear (3). As no effective treatment for OLK exists in clinical practice (4,5), studying the mechanism and determining new potential molecular markers of OLK and effective therapeutic targets is important.
Circular RNAs (circRNAs) are newly identified non-coding RNAs with a covalently closed loop structure with no 5' or 3' polarities or polyadenylated tails (6). It was once thought that circRNAs were byproducts of intramolecular mis-splicing (7). They are considered to be a new class of widespread, highly abundant, conserved, and stable endogenous non-coding RNAs in eukaryotic cells (8-10). With the development of bioinformatic approaches, biochemical enrichment strategies, and high-throughput sequencing technology, a number of circRNAs have been defined in various species and diseases.
CircRNAs can participate in and regulate human diseases at the epigenetic level via competing for endogenous RNA (ceRNA) mechanisms. One hypothesis proposed that transcripts might use micro RNA (miRNA) response elements (MREs) to form a large-scale regulatory network across the transcriptome (11). The hypothesis can be interpreted to posit that any RNA transcript with MREs may serve as a ceRNA. Compared with other types of ceRNAs, circRNAs are highly expressed and stable in cells and have more binding sites with miRNAs, which enables effective regulation of miRNAs (12,13).
The functions of circRNAs suggest that they have a vital role in disease regulation (14,15). Studies on the relationship between circRNAs and diseases have included cancers, and nervous system and cardiovascular diseases (16-18). Many studies have shown that circRNAs can be used as biomarkers and therapeutic targets (19-21). Several studies have reported on the role of circRNAs in disease progression in the field of stomatology, such as in oral squamous cell carcinoma (OSCC) and oral mucosal melanoma (OMM) (22,23). Research indicates that circRNA_100290, circular RNA DOCK1 (circDOCK1), and circ_0007059 are involved in the regulation of growth and metastasis in OSCC through the mechanism of ceRNA (24-26). Tumorigenesis and metastasis of OMM can be regulated by hsa_circ_0005320, hsa_circ_0067531, and hsa_circ_0000869 (27). All of these findings indicate that circRNA may play a part in oral cancer, and provide fresh motivation for identifying whether circRNAs participate in the regulation of oral precancerous lesion.
Accordingly, we explored the expression profiles of circRNAs in OLK and normal oral mucosal (NOM) tissue by high-throughput RNA sequencing, and analyzed the screened, differentially expressed (DE) genes. We generated the miRNA-mRNA and ceRNA networks. It was confirmed that circHLA-C is a potential biomarker for diagnosis and prognosis and that it might promote the development of OLK.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/atm-20-3840).
Methods
Tissue specimens
Six OLK tissue samples were obtained from two male and four female patients who received treatment at the Department of Oral Mucosa of the Shanghai Ninth People’s Hospital, from September 2018 to November 2018 (Table 1). Based on the diagnostic criteria of World Health Organization, the patients were diagnosed by histopathological examination. Six NOM specimens were prospectively obtained from the Department of Plastic and Reconstructive Surgery. None of these selected objects had the systemic disease, a history of immune stimulants (including corticosteroid) within 3 months and radiotherapy and chemotherapy. All tissue specimens were frozen in liquid nitrogen immediately. Before surgery, we provided a written informed consent for all patients for experimental use of their tissues. Specimen information was recorded and data were encoded to protect patient privacy. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) individual consent for this retrospective analysis was waived. All the studies were approved by the Ethics Committee of Shanghai Ninth People’s Hospital [No. (2012) 21].

Full table
RNA extraction and quality control
Total RNA was extracted using TRIzol reagent (Life Technologies, Carlsbad, CA, USA). The RNA concentrations for each specimen were measured using a NanoDrop ND-1000 (Thermo Fisher Scientific, Waltham, MA, USA). Spectrophotometer OD260/OD280 values were used for RNA purity indexes. Quality control results indicated a range of OD260/OD280 between 1.8 and 2.1. Sufficient RNA was used with Ribo-Zero rRNA Removal Kits (Illumina, USA) to eliminate ribosomal RNA (rRNA) according to kit instructions. RNA libraries were constructed using rRNA-depleted RNAs and TruSeq Stranded Total RNA Library Prep Kits (Illumina, USA). RNA libraries were adjusted to 10 pM and converted into denatured single-stranded DNA molecules and amplified in situ as hierarchical clusters. Sequencing libraries were detected by an Agilent 2100 Bioanalyzer using an Agilent DNA 1000 chip kit (Agilent, Technologies, USA).
CircRNA sequencing data analysis
Paired-end reads were harvested from Illumina HiSeq 4000 sequencer, and were quality controlled by Q30. After 3’ adaptor-trimming and low-quality reads removing by cutadapt software (v1.9.3), screened and qualified trimmed reads were aligned to a reference genome or transcriptome with STAR software (v2.5.1b). Detection and identification were performed by DCC software (v0.4.4). The identified circRNAs were annotated using circBase and circ2Traits databases (28,29). According to sequencing the depth and degree of variation, we normalized the data and screened for DE circRNAs between OLK and NOM using edge R software (v3.16.5). To generate the profiling of DE circRNAs between OLK and NOM, hierarchical clustering analysis was performed based on the expression levels of all identified circRNAs, and the significant difference between OLK and NOM was analyzed by Cluster and TreeView software (30).
Unprocessed and analyzed sequencing data, after standardization, were uploaded to the National Center for Biotechnology Information Gene Expression Omnibus (GEO). The approved GEO accession number is GSE131568.
Quantitative real-time RT–PCR
According to the results of the circRNAs expression and functional analysis, 10 significantly altered circRNAs were selected for real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) using the 6 pairs of samples. Tissues of OLK and NOM from each patient were considered as an independent cohort to verify the candidate circHLA-C by qRT-PCR. The independent cohort contained samples from 40 patients, whose data are shown in Table S1.
SuperScript III reverse transcription kits (Invitrogen, Carlsbad, USA) were used according to the manufacturer’s instructions to construct cDNA libraries of extracted RNA. qRT-PCR was conducted by ViiA 7 Real-time System (Applied Biosystems, Wilmington, DE, USA) using qPCR SYBR Green master mix (Cloudseq, Shanghai, China). Divergent primer pairs designed for target 10 circRNAs selected were summarized in Table 2 (Sangon, Shanghai, China). β-actin was the internal control. Target circRNAs and β-actin were amplified routinely in triplicate. Relative expression of circRNAs was expressed as 2−∆∆Ct of each circRNA measurement.
Full table
Construction of ceRNA network and bioinformatic analysis
Potential target miRNAs of circRNAs were predicted by TargetScan. The bioinformatics software Cytoscape (v2.8.0) was used to construct the network of circRNAs and their downstream miRNAs and mRNAs.
The predicted functions of the DE circRNAs between OLK and the NOM control were determined by performing Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. The GO covers three domains: biological process (BP), cellular component (CC), and molecular function (MF). The terms of GO were screened on the basis of host genes with significantly different circRNAs and the GO annotation list. The P value denoted the significance of GO term enrichment in the host genes using Fisher’s exact test (P<0.05 was recommended). The KEGG analysis was used for annotated host genes with DE circRNAs using the KEGG database. The Fisher P value denoted the significance of each pathway involved, and P<0.05 was deemed to indicate statistically significant differences.
Statistical analysis
Statistical software SPSS 19.0 (SPSS, Chicago, IL, USA) and Graphpad Prism version 8.0 (Grapahpad, San Diego, CA, USA) were used for management and statistical analysis of the data. Statistical significance of differences between two groups was performed by unpaired t-test (*, P<0.05, **, P<0.01, ***, P<0.001, ****, P<0.0001). Quantitative data was expressed as mean ± SEM (standard error of mean). All the experiments were performed three times at least. ROC curve analysis was utilized to identify the sensitivity and specificity of circHLA-C. Correlation between circHLA-C and epithelial dysplasia was analyzed by Spearman’s rank correlation coefficient.
Results
Profiling and characteristics of circRNAs in human OLK and NOM
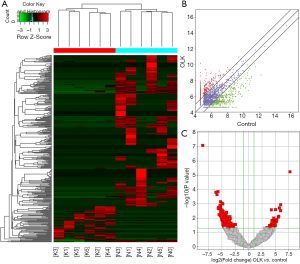
The differentiated circRNAs between OLK and NOM groups were compared in 6 pairs of the OLK and NOM samples. In contrast to the NOM group, hierarchical clustering diagrams predicted relationships among the samples (Figure 1A). Scatter and volcanic plots were painted with statistical criteria that had been determined through fold change (FC) and P value (Figure 1B,C). In total, 366 significantly dysregulated circRNAs were identified in OLK, of which 65 were markedly upregulated and 301 downregulated.
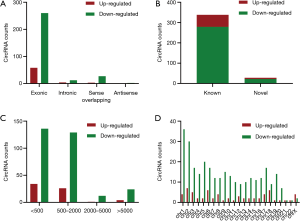
Among the 366 dysregulated circRNAs, 28 new circRNAs were identified, with the remainder already existing in the circbase (28). Most were exonic and known circRNAs (Figure 2A,B). The lengths of these circRNAs were concentrated into two groups: <500 and 500–2,000 nucleotides (Figure 2C). The circRNAs were located on nearly all human chromosomes, including the 22 autosomes and X chromosomes (Figure 2D).
Further validation of the selected circRNA
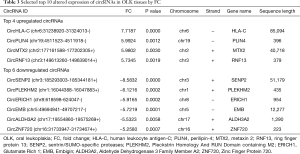
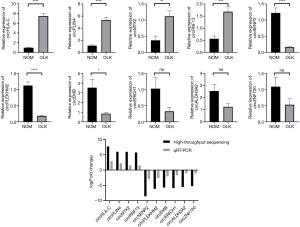
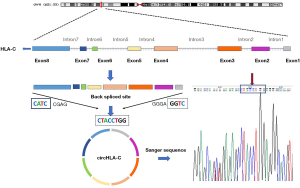
The top 4 upregulated and 6 downregulated circRNAs were selected for verification by qRT-PCR (Table 3). Validation was performed on the 6 pairs of tissue. Of the 10 circRNAs, 7 showed the same change in direction and statistical significance as the circRNA sequencing (Figure 3). Since circRNA is not sensitive to nuclease, we selected 5 interesting circRNAs from 7 circRNAs verified by qRT-PCR for enzyme tolerance testing. The results showed that circHLA-C, circPLIN4 (perilipin-4), and circRNF13 (ring finger protein 13) could resist the hydrolysis of ribonuclease (RNase) R. Then Sanger sequencing was conducted on the three circRNAs. The experiment results revealed that the most upregulated circRNA (circHLA-C) showed that it had a specific back-splice junction site. We further performed a visualization analysis of circHLA-C, which was derived from HLA-C and consisted of 8 exons and 7 introns. The back-splice site was CTACCTGG, as verified by Sanger sequencing (Figure 4).
Full table
Predicted function of DE circRNAs in OLK
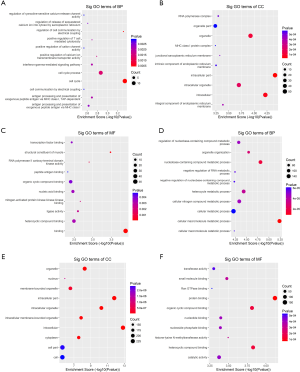
The functions of DE circRNAs in OLK tissues were predicted because of the ability of circRNA to promote the expression of source genes. The GO enrichment analysis was based on a tripartite functional analysis of circRNA host genes that BP, CC, and MF (Figure 5A,B,C,D,E,F). In upregulated DE circRNAs, HLA-C, the host gene of circHLA-C, participated in as many as 95 GO terms, including 44 BP, 45 CC, and 6 MF, respectively. Multiterm of T cell-mediated cytotoxicity and immunity was regulated by HLA-C in BP. Major histocompatibility complex (MHC) class I protein was regulated by HLA-C in CC.
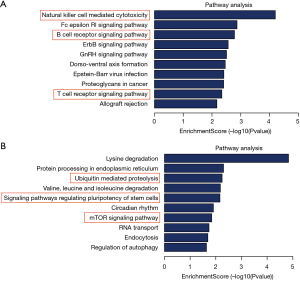
The KEGG analysis identified functional pathways through accurate pathways relevant to their source genes, with 139 KEGG pathways associated with 366 DE genes (DEGs). As a precancerous lesion, OLK is involved in the process of carcinogenesis through 3 pathways of the KEGG pathway. Among 60 upregulated circRNA pathways, “proteoglycans in cancer,” “miRNAs in cancer,” and “pathways in cancer” are all involved in regulating the expression of extracellular-signal-regulated protein kinase/mitogen-activated protein kinase (ERK/MAPK1), which have been proved to be crucial factors in the pathogenesis of OLK (31,32). This suggests that OLK is a risk of oral cancer on account of it being involved in several cancer-related pathways. Moreover, we found that the regulation of the “natural killer cell-mediated cytotoxicity,” “T cell receptor signaling pathway,” and “B cell receptor signaling pathway” were all related to immunity, and were significantly regulated by the 65 overexpressed circRNAs. We also found that circHLA-C was regulated in the pathway of “natural killer cell-mediated cytotoxicity”, a detailed mechanism diagram is shown in Figure S1. Among 79 downregulated circRNA pathways, “ubiquitin-mediated proteolysis”, “signaling pathways regulating pluripotency of stem cells”, and “mTOR signaling pathway” were significantly regulated by the 301 downregulated circRNAs (Figure 6A,B).
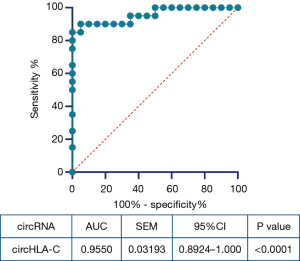
ROC curve analysis of confirmed diagnosability of circHLA-C in OLK
The ROC curve analysis was performed to evaluate the potential diagnosability of circHLA-C. Among another 20 pairs of OLK and NOM, qRT-PCR was conducted to confirm the prognostic value. The area under the curve (AUC) was found for circHLA-C (AUC 0.9550, 95% CI, 0.8924–1.000, P value <0.0001). Generally speaking, the larger the AUC, the stronger the diagnosability. We foresee that circHLA-C may potentially function as a valuable biomarker for the diagnosis of OLK (Figure 7).
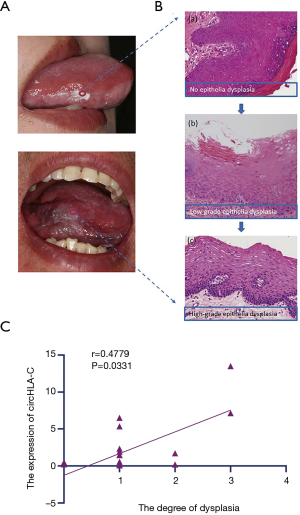
Correlation between circHLA-C and epithelia dysplasia
It was observed that circHLA-C was expressed differently in different degrees of dysplasia of OLK tissues. According to pathological results, dysplasia of OLK could be divided into three levels, being mild, moderate, and severe (Figure 8A,B). The correlation between the expression of circHLA-C and the degree of dysplasia in another 20 samples was analyzed. The result delineated that their expression showed a positive correlation, with a statistically significant difference between the expression of circHLA-C and the mild and moderate degree of OLK dysplasia (P<0.05) (Figure 8C).
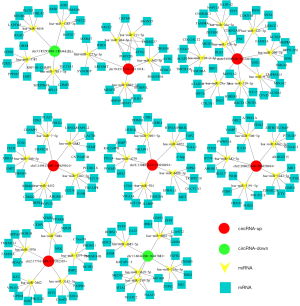
CircRNA/miRNA interaction analysis
One way that circRNAs can regulate miRNA levels is by absorbing miRNA and acting as a “miRNA sponge” (12). We screened the top 8 circRNAs and established a ceRNA network. The top 5 miRNAs regulated by each circRNA are listed in Figure 9. It was predicted that circRNAs such as circRNF13 (chr3:149613260-149639014+) could adsorb miR-7-5p, miR-5683, miR-7107-3p, miR-4452, and miR-580-5p. Bioinformatics prediction continued to analyze the downstream of the miRNA-mRNA network. The top 5 mRNAs that miRNA could inhibit in the downstream were also predicted, which highlighted that the roles of circRNA are very extensive, and can be regulated by several different pathways (Figure 9).
Discussion
OSCC is the sixth most common malignant tumor in the world; however, patients are often diagnosed at the advanced stage when the prognosis is poor. On average, cancer takes four to five years to develop from OLK into OSCC (33,34); hence it is very important for diagnosis and the initiation of treatment to occur in a well-established premalignant stage. However, for a considerable number of OLKs, the pathogenesis remains ambiguous, and early diagnosis and prognosis biomarkers for OLK are not commonly applied in laboratory and clinical settings. Therefore, it is vital to provide a solution to fill this gap and to discover effective prevention and treatment strategies for OLK by researching new biomarkers.
This study proposed the concept of miRNA sponges, with circRNAs acting as antagonists of miRNAs, as it has been observed with the circRNA CDR1as (6). In addition, some studies have suggested that circRNAs regulate the expression of their parental genes in cis and promote gene transcription (35). Other studies showed that circRNAs bind to proteins to affect cell cycle progression and prevent protein function (36). In this study, the important functions of how circRNA worked in the occurrence and development of OLK were discussed and predicted.
We know that circRNA is a non-coding RNA that plays an important role in disease mechanisms (37,38). A large number of circRNAs have been identified in various cell lines and species (13,38,39). In response to there being no relevant literature about this topic, we studied the circRNA expression profiles for OLK. Some studies suggest that circRNAs regulate the expression of their parental genes in cis and promote gene transcription (35). We found that circHLA-C was derived from HLA-C, the human MHC, namely human leukocyte antigen. The efficacy of the immune response to infectious and autoimmune diseases can be influenced by variations in the expression of HLA-C. The evolution of HLA-C has led to it having more intensive interactions with killer cell immunoglobulin-like receptors (KIRs), and thus it can influence the regulation of NK cell responses (40). Studies show that the genotype of KIR2DL1(+)-HLA-C2(+) could serve as a risk factor in predisposing people to OSCC at a younger age (41). In addition, variation in HLA genes may impact the risk of squamous cell cervical cancer by altering the efficiency of the T-cell-mediated immune response to human papillomavirus (HPV) antigens (42).
The GO enrichment analysis showed that DE genes were involved in a number of biological processes that could regulate the occurrence and development of diseases by influencing biological processes, cell components, and MFs. According to GO enrichment scores, “cell cycle”, “positive regulation of T cell-mediated cytotoxicity”, and “antigen procession and presentation of exogenous peptide antigen via major histocompatibility (MHC) class I molecules” were the top GO terms in BP. Most DEGs were focused on “cell cycle” and “cell cycle process” because epithelial dysplasia is one of the typical pathological manifestations of OLK, and it causes alterations in the cell cycle. It is speculated that changes in the expression of genes that control the cell cycle are the main causes of OLK. “T cell-mediated cytotoxicity and immunity” and “MHC class I protein complex” are regulated by HLA-C. Studies have shown that MHC class I molecules are connected with recognition by specific T‐cell receptors and molecules present on the surface of CD8-positive (CD8+) cells (43). It was reported that MHC class I, a pivotal etiological factor, when reduced in OLK, is related to the degree of dysplasia (44,45). Moreover, it is known that CD8+ cells were significantly more abundant in OSCC and OLK with dysplasia compared with OLK without dysplasia (46). It was indicated that the abnormal expression of key genes that regulate T cell-mediated cytotoxicity, and antigen procession and presentation of exogenous peptide antigen via MHC class I molecules could function sustainedly in the course of occurrence and dysplasia of OLK. As the gene with the most significant expression difference, circHLA-C may be an important part of DE genes in this process.
The KEGG pathway analysis predicted 139 pathways related to DE circRNAs. Among 60 upregulated circRNA pathways, the top 10 pathways included “natural killer cell-mediated cytotoxicity”, “B cell receptor signaling pathway”, “Epstein-Barr virus infection”, “proteoglycans in cancer”, “T cell receptor signaling pathway”, and “allograft rejection”. Among these pathways, we found that the target gene of circHLA-C is involved in the top 3 pathway “natural killer cell-mediated cytotoxicity”, “Epstein-Barr virus infection”, and “allograft rejection”. This result suggested that OLK may be associated with infection and immunosuppressive factors, and circHLA-C may act as a key figure. These important pathways are of great significance in OLK development and will guide the discussion regarding the mechanisms of OLK in the future. The mechanism of circHLA-C’s participation in the pathway still needs to be verified by subsequent experiments.
We further validated that circHLA-C was significantly increased in repeated OLK samples of 20 OLK patients as compared with healthy samples. The ROC curve showed that circHLA-C had a satisfactory AUC. It was not only indicated that overexpressed circHLA-C plays an important role in the pathogenesis of OLK, but also demonstrated that circHLA-C might define its clinical value as a novel biomarker.
Studies have shown that circRNA can be used as ceRNA to adsorb miRNA through a sponge mechanism and act on a cellular regulatory network to establish a large gene regulatory transcriptome network using small RNA (47). The ceRNA network we constructed showed that circHLA-C could perform complementary pairing with miR-9, miR-339-5p, miR-29a-3p, miR-26a-5p, and miR-222-3p through the sponge mechanism because perfect binding sites were predicted between circHLA-C and above miRNA via TargetScan software. According to literature reports, the expression of serum miR-9 was significantly reduced in patients with OLK and OSCC compared with NOM samples (48). Downregulation of miR-339-5p was reported in OLK and some primary cancer tissues as a tumor suppressor and had been associated with cancer progression (49,50). Significant downregulation of miR-29a-3p and miR-26a-5p was found in OLK and cancer tissues; this downregulation implied a tumor-suppressive function (51). It was also found that miR-222-3p was significantly downregulated in OLK compared to normal and OSCC patients (52). As we can see, a circRNA may have multiple binding sites of the same miRNA or multiple miRNA binding sites. It might be that circHLA-C sponges these miRNAs to regulate the initiation and progression of OLK. More specific research should be conducted on the interaction between circHLA-C and miRNAs and the downstream target genes in OLK in the future. Time-specificity is a property of circRNA, with the expression of circRNA changing at different stages of cell development (9). During the transformation of OLK to OSCC, the degree of dysplasia of cells gradually increases, which could reflect the severity of OLK, leading us to ponder whether the process is the result of circRNA regulation. In the results, the difference of circHLA-C expression in different tissues made us contemplate whether it was correlated with the degree of dysplasia of OLK samples. The results showed that the expression of circHLA-C correlated positively with the degree of the dysplasia (with no dysplasia to moderate dysplasia) (P=0.01589). However, our samples were lacking in cases of severe dysplasia; and an increase in sample size and degrees of dysplasia is needed for further validation. Further cell and animal model experiments should be undertaken to better explain the details of the mechanism and specific functions of circHLA-C in OLK.
Conclusions
Our study is the first to show that a large number of circRNAs were significantly DE between NOM and OLK tissues. The relationship between circHLA-C and dysplasia indicated that circHLA-C may positively correlate with the degree of dysplasia and plays a pivotal role in the immunosuppression and infection disorder of OLK. Contrary correlation and predicted binding sites between circHLA-C and miR-9, miR-339-5p, miR-29a-3p, miR-26a-5p, and miR-222-3p suggested that circHLA-C may regulate the progress of OLK via sponging miRNA from above. In summary, circHLA-C may be a valuable biomarker in OLK diagnosis, and it has generated new inspiration for future research into the diagnosis and prognosis of OLK.
Acknowledgments
We would like to thank Professor DJC and Professor YPP for providing NOM tissues and CloudSeq Biotech (Shanghai, China) for assistance with high-throughput sequencing service.
Funding: This work was supported by National Key R&D Program of China (2017YFC0840100, 2017YFC0840110).
Footnote
Reporting Checklist: We present the following article in accordance with the MDAR checklist. Available at http://dx.doi.org/10.21037/atm-20-3840
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3840
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-3840
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3840). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Shanghai Ninth People’s Hospital [No.: 2012 (21)] and individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Axéll T, Pindborg JJ, Smith CJ, et al. Oral white lesions with special reference to precancerous and tobacco- related lesions: conclusions of an international symposium held in Uppsala, Sweden, May 18-21 1994. International Collaborative Group on Oral White Lesions. J Oral Pathol Med 1996;25:49-54. [Crossref] [PubMed]
- Hong WK, Endicott J, Itri LM, et al. 13-cis-retinoic acid in the treatment of oral leukoplakia. N Engl J Med 1986;315:1501-5. [Crossref] [PubMed]
- Lee JJ, Hong WK, Hittelman WN, et al. Predicting cancer development in oral leukoplakia: ten years of translational research. Clin Cancer Res 2000;6:1702-10. [PubMed]
- van der Waal I, Schepman KP, van der Meij EH, et al. Oral leukoplakia: a clinicopathological review. Oral Oncol 1997;33:291-301. [Crossref] [PubMed]
- Warnakulasuriya S, Ariyawardana A. Malignant transformation of oral leukoplakia: a systematic review of observational studies. J Oral Pathol Med 2016;45:155-66. [Crossref] [PubMed]
- Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495:333-8. [Crossref] [PubMed]
- Cocquerelle C, Mascrez B, Hétuin D, et al. Mis-splicing yields circular RNA molecules. FASEB J 1993;7:155-60. [Crossref] [PubMed]
- Rybak-Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 2015;58:870-85. [Crossref] [PubMed]
- Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016;7:11215. [Crossref] [PubMed]
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014;32:453-61. [Crossref] [PubMed]
- Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 2011;146:353-8. [Crossref] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495:384. [Crossref] [PubMed]
- Guo JU, Agarwal V, Guo H, et al. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014;15:409. [Crossref] [PubMed]
- Bach DH, Lee SK, Sood AK. Circular RNAs in Cancer. Mol Ther Nucleic Acids 2019;16:118-29. [Crossref] [PubMed]
- Altesha MA, Ni T, Khan A, et al. Circular RNA in cardiovascular disease. J Cell Physiol 2019;234:5588-600. [Crossref] [PubMed]
- Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer 2018;17:79. [Crossref] [PubMed]
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20:675-91. [Crossref] [PubMed]
- Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer 2019;18:20. [Crossref] [PubMed]
- Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell 2018;71:428-42. [Crossref] [PubMed]
- Wilusz JE. A. 360° view of circular RNAs: From biogenesis to functions. Wiley Interdiscip Rev RNA 2018;9:e1478. [Crossref] [PubMed]
- Chen B, Huang S. Circular RNA: An emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett 2018;418:41-50. [Crossref] [PubMed]
- Wang YF, Li BW, Sun S, et al. Circular RNA Expression in Oral Squamous Cell Carcinoma. Front Oncol 2018;8:398. [Crossref] [PubMed]
- Qiu X, Ke X, Ma H, et al. Profiling and bioinformatics analyses reveal differential expression of circular RNA in tongue cancer revealed by high-throughput sequencing. J Cell Biochem 2019;120:4102-12. [Crossref] [PubMed]
- Chen X, Yu J, Tian H, et al. Circle RNA hsa_circRNA_100290 serves as a ceRNA for miR-378a to regulate oral squamous cell carcinoma cells growth via Glucose transporter-1 (GLUT1) and glycolysis. J Cell Physiol 2019;234:19130-40. [Crossref] [PubMed]
- Wei Y, Yan Y, Wang H, et al. CircDOCK1 suppresses cell apoptosis via inhibition of miR-196a-5p by targeting BIRC3 in OSCC. Oncol Lett 2018;39:951-66.
- Su W, Wang Y, Wang F, et al. Circular RNA hsa_circ_0007059 indicates prognosis and influences malignant behavior via AKT/mTOR in oral squamous cell carcinoma. J Cell Physiol 2019;234:15156-66. [Crossref] [PubMed]
- Ju H, Zhang L, Mao L, et al. Altered expression pattern of circular RNAs in metastatic oral mucosal melanoma. Am J Cancer Res 2018;8:1788-800. [PubMed]
- Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA 2014;20:1666-70. [Crossref] [PubMed]
- Ghosal S, Das S, Sen R, et al. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet 2013;4:283. [Crossref] [PubMed]
- Saldanha AJ. Java Treeview-extensible visualization of microarray data. Bioinformatics 2004;20:3246-8. [Crossref] [PubMed]
- Li Z, Gao M, Liu C, et al. Immunohistochemical Detection of Aurora A and ERK Pathway in Oral Leukoplakia and Oral Squamous Cell Carcinoma. J Hard Tissue Biol 2014;23:71-6. [Crossref]
- Yang W, Wang XM, Yuan HY, et al. Exploring the mechanism of WWOX growth inhibitory effects on oral squamous cell carcinoma. Oncol Lett 2017;13:3198-204. [Crossref] [PubMed]
- Neville BW, Day TA. Oral cancer and precancerous lesions. CA Cancer J Clin 2002;52:195-215. [Crossref] [PubMed]
- Brouns E, Baart J, Karagozoglu Kh, et al. Malignant transformation of oral leukoplakia in a well-defined cohort of 144 patients. Oral Dis 2014;20:e19-e24. [Crossref] [PubMed]
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015;22:256-64. [Crossref] [PubMed]
- Du DD, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 2016;44:2846-58. [Crossref] [PubMed]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol Cell 2014;56:55-66. [Crossref] [PubMed]
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013;19:141-57. [Crossref] [PubMed]
- Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 2016;17:205-11. [Crossref] [PubMed]
- Kaur G, Gras S, Mobbs JI, et al. Structural and regulatory diversity shape HLA-C protein expression levels. Nat Commun 2017;8:15924. [Crossref] [PubMed]
- Dutta A, Saikia N, Phookan J, et al. Association of killer cell immunoglobulin-like receptor gene 2DL1 and its HLA-C2 ligand with family history of cancer in oral squamous cell carcinoma. Immunogenetics 2014;66:439-48. [Crossref] [PubMed]
- Madeleine MM, Johnson LG, Smith AG, et al. Comprehensive analysis of HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 loci and squamous cell cervical cancer risk. Cancer Res 2008;68:3532-9. [Crossref] [PubMed]
- Goldszmid RS, Idoyaga J, Bravo AI, et al. Dendritic cells charged with apoptotic tumor cells induce long-lived protective CD4+ and CD8+ T cell immunity against B16 melanoma. J Immunol 2003;171:5940-7. [Crossref] [PubMed]
- Wu PF, Han BF, Xia H, et al. The expression of human major histocompatibility complex-I in oral leukoplakia. Hua Xi Kou Qiang Yi Xue Za Zhi 2010;28:439-42. [PubMed]
- Prime SS, Pitigala-Arachchi A, Crane IJ, et al. The expression of cell surface MHC class I heavy and light chain molecules in pre-malignant and malignant lesions of the oral mucosa. Histopathology 1987;11:81-91. [PubMed]
- Öhman J, Magnusson B, Telemo E, et al. Langerhans cells and T cells sense cell dysplasia in oral leukoplakias and oral squamous cell carcinomas--evidence for immunosurveillance. Scand J Immunol 2012;76:39-48. [Crossref] [PubMed]
- Liang Y, Zhang YZ, Xu LA, et al. CircRNA Expression Pattern and ceRNA and miRNA-mRNA Networks Involved in Anther Development in the CMS Line of Brassica campestris. Int J Mol Sci 2019;20:4808. [Crossref] [PubMed]
- Sun L, Liu L, Fu H, et al. Association of Decreased Expression of Serum miR-9 with Poor Prognosis of Oral Squamous Cell Carcinoma Patients. Med Sci Monit 2016;22:289-94. [Crossref] [PubMed]
- Maimaiti A, Abudoukeremu K, Tie L, et al. MicroRNA expression profiling and functional annotation analysis of their targets associated with the malignant transformation of oral leukoplakia. Gene 2015;558:271-7. [Crossref] [PubMed]
- Zhang L, Lu XQ, Zhou XQ, et al. NEAT1 induces osteosarcoma development by modulating the miR-339-5p/TGF-β1 pathway. J Cell Physiol 2019;234:5097-105. [Crossref] [PubMed]
- Roy R, Singh R, Chattopadhyay E, et al. MicroRNA and target gene expression based clustering of oral cancer, precancer and normal tissues. Gene 2016;593:58-63. [Crossref] [PubMed]
- Chang YA, Weng SL, Yang SF, et al. A Three-MicroRNA Signature as a Potential Biomarker for the Early Detection of Oral Cancer. Int J Mol Sci 2018;19:758. [Crossref] [PubMed]


