
Weighted gene co-expression network analysis identified underlying hub genes and mechanisms in the occurrence and development of viral myocarditis
Introduction
Myocarditis is an inflammatory myocardial disease, which is mainly caused by viral infections, such as those caused by coxsackieviruses, adenoviruses, influenza viruses, cytomegaloviruses, and human immunodeficiency virus (1). Pathologically, myocarditis is characterised by infiltration of the myocardium with mononuclear cells (2). Most myocarditis patients exhibit mild symptoms. However, a subset of patients experience fulminant myocarditis, leading to heart failure, arrhythmia, fulminant haemodynamic collapse and sudden mortality (3-5), whereas about one third of viral myocarditis patients develop a chronic form of the disease, which may lead to dilated cardiomyopathy (DCM) (1).
As we know, direct injury caused by the virus and indirect injury mediated by the host’s immune response underlie the pathogenesis of viral myocarditis (6). The progression of viral myocarditis is characterised by 3 stages: (I) an acute stage triggered by viral entry and replication, (II) a subacute stage characterised by inflammatory cell infiltration, and (III) a chronic stage characterised by cardiac remodelling (7). However, the mechanism underlying fulminant myocarditis and how it progresses into dilated cardiomyopathy still require further elucidation.
Conventional molecular biology techniques can uncover the specific function of a gene, but do not reveal interaction between genes in biological networks (8,9). To our knowledge, there is still a lack of systematic biological network analyses of hub genes in viral myocarditis. Weighted gene co-expression network analysis (WGCNA), a widely used systems biology approach, is a powerful tool that is used to define correlation patterns between genes (10-15). Here, we constructed a co-expression network using expression data from the Gene Expression Omnibus (GEO) database. Genes with similar expression patterns were clustered into the same modules. The relationships between different stages of the myocarditis and modules were calculated to identify highly related modules. The turquoise module was positively correlated with the acute disease stage, while the brown and yellow modules were positively and negatively correlated with the chronic disease stage, respectively. The hub genes and miRNAs identified in these modules could serve as biomarkers and therapeutic targets for viral myocarditis.
Methods
Data collection
The heart RNA expression profiles from mouse models of viral myocarditis were downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/; GSE35182) (16). This dataset was generated on the GPL6246 platform of the Affymetrix Mouse Gene 1.0 ST Array. To explore the occurrence and development of viral myocarditis, we analysed the RNA expression profiles in an acute infection group (coxsackievirus B3 infection for 10 days), chronic infection group (coxsackievirus B3 infection for 90 days) and normal group (PBS injection).
Data pre-processing
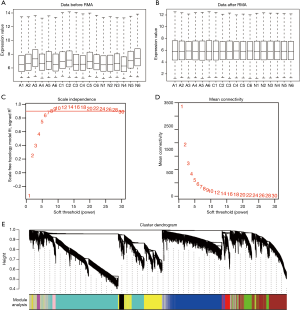
The R version 3.6.1 software was used for data processing and analysis. The raw data in CEL format were read using the Affy package version 1.62.0 (17) and processed using the robust multiarray average (RMA) algorithm (Figure 1A,B) (18). Next, we used the impute.knn function in the impute package version 1.58.0 to impute the individual missing values in the raw data. This function used the Euclidean metric to find k nearest genes that were similar to the expression profiles of each gene with a missing value, and estimated the missing value through the expression values of nearest genes (19,20). After processing, the 6,680 genes exhibiting the most significant expression changes (the top 25% of rank genes with the largest variance) were selected for further analysis.
Co-expression network construction
The WGCNA package version 1.68 in R software was used to construct the co-expression network (21,22). An appropriate soft threshold was selected to ensure scale-free topology (R2>0.9). The topological overlap matrix (TOM) was constructed to measure the network connectivity of the genes. Genes with similar patterns were clustered into the same modules (minimum size =30) using average linkage hierarchical clustering. The relationships between phenotypes and modules were calculated to identify highly related modules. Finally, the highly correlated modules were analysed to explore their potential roles. In addition, the gene expression profiles of the highly correlated modules were visualised using the R software.
Functional annotation
The functional annotation analyses of the highly correlated modules were done using the clusterProfiler package version 3.12.0 on R (23,24). The P value was adjusted by the Holm–Bonferroni method. The adjusted P value cutoff was set at 0.05 and the q-value cutoff was set at 0.2. An adjusted P value less than 0.05 was considered to be significant, and the identified significant analyses were sorted by gene counts.
Identification of hub genes
Hub genes are defined as the genes that exhibit the greatest association with a disease. The IC for each gene was calculated by summing the connection strengths with other genes in the same module. For a given gene, the higher the IC, the stronger its relationship with other genes, and the more important it is within the module. Therefore, hub genes are the genes that possess high IC values. The interaction networks between hub genes and other genes in the module were visualised using Cytoscape version 3.7.2 (25).
Prediction of potential miRNA targeting networks
The top 100 genes ranked by IC were selected from each module and a miRNA targeting network was predicted by miRNet analysis (www.mirnet.ca) (18). The network was visualised using Cytoscape version 3.7.2.
Statistical analysis
WGCNA and functional annotation were performed in R (version 3.6.1) with default test statistics and cutoff values as specified in individual Methods sections. P<0.05 was considered statistically significant.
Results
Construction of weighted co-expression network and identification of key modules
We used the 6,680 genes that exhibited the greatest differences in expression to construct weighted gene co-expression networks. A total of 18 samples were used as input for the hierarchical clustering analysis. The hclust function was then used to cluster the samples and visualise outliers (18). Because this analysis did not reveal any outliers, all the samples were included in the downstream analyses (Figure S1). Before constructing the weighted co-expression matrix, a soft-threshold β was calculated to ensure a scale-free topology. When the soft-threshold β was equal to 14 the independence degree rose to 0.964. Therefore, co-expression gene modules were constructed using β=14 (Figure 1C,D).
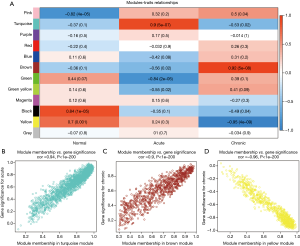
Next, the co-expression network was constructed using a one-step method and the correlation matrix was constructed to calculate the correlation efficiency between genes. Genes with similar expression patterns were clustered into the same module. A total of 12 modules were identified (Figure 1E). Next, module eigengenes (MEs), components of a module representative of the gene expression profiles in that module, were calculated for each module. Subsequently, the correlation between MEs and viral myocarditis at different stages was calculated, and a correlation coefficient greater than, or equal to, 0.9 or less than, or equal to, −0.9 (| r | ≥0.9) was considered to be significant. In acute viral myocarditis, the turquoise module (r=0.9; P=5×10−7) was identified as the statistically significant module. In chronic viral myocarditis, the brown (r=0.92; P=5×10−8) and yellow (r=−0.95; P=4×10−9) modules were identified as the statistically significant modules (Figure 2A). These modules were then selected for further analyses. To evaluate the correlation between the different stages of viral myocarditis and the statistically significant modules, we calculated the values between module membership (MM) and gene significance (GS). The turquoise module (cor =0.94; P<1×10−200) showed a high positive correlation with acute viral myocarditis (Figure 2B), whereas the brown module (cor =0.9; P<1×10−200) showed a high positive correlation with chronic viral myocarditis. A high negative correlation (cor =−0.96; P<1×10−200) was observed between the yellow module and the chronic disease stage (Figure 2C,D).
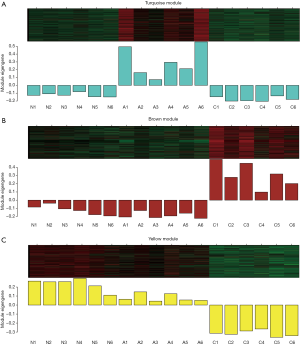
Genes expression of key modules at the different stages of viral myocarditis
To further explore the role of key modules in the occurrence and development of viral myocarditis, the gene expression in these modules at the different stages of viral myocarditis were visualised using a heatmap. Gene expression in the turquoise module was significantly increased in the acute infection group (Figure 3A), indicating that genes in this module play an important role in the acute stage of viral myocarditis. In chronic viral myocarditis, gene expression in the brown module was significantly increased, suggesting that the genes in this module play an important role in chronic viral myocarditis (Figure 3B). In the yellow module, gene expression continued to fall reaching its lowest level in the chronic phase, indicating that genes in this module are important for the development of viral myocarditis (Figure 3C).
Functional annotation of the key modules
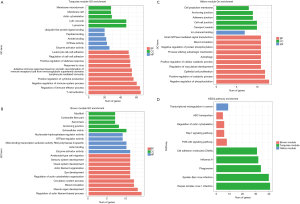
As the turquoise, brown and yellow modules were identified as key modules, functional annotation was adopted to characterise them. Gene Ontology (GO) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were executed using clusterProfiler. To avoid inaccurate enrichment results caused by too many genes, we selected the top 25% genes ranked by IC in each module for enrichment analyses.
The functional enrichment of GO term categories includes biological process (BP), cellular component (CC), and molecular function (MF). Analysis of BP in the turquoise module revealed that the genes were enriched in T cell activation, regulation of immune effector processes, negative regulation of immune system processes, positive regulation of cytokine production, and lymphocyte mediated immunity. Regarding the CC and MF categories, the genes were mainly associated with the lysosome (CC), lytic vacuole (CC), enzyme activator activity (MF) and GTPase activity (MF) (Figure 4A). The most enriched BPs in the brown module were regulation of actin filament-based processes, muscle organ development, blood circulation, circulatory system process, and regulation of actin cytoskeleton organization. In the CC and MF categories, genes were mainly associated with the extracellular matrix (CC), anchoring junction (CC), enzyme activator activity (MF), and actin binding (MF) (Figure 4B). The most enriched BPs in the yellow module were negative regulation of phosphorylation, positive regulation of catabolic process, epithelial cell proliferation, regulation of vasculature development, and autophagy. In the CC and MF categories, genes were mainly associated with transport vesicles (CC), cell-cell junction (CC), and ion channel binding (MF) (Figure 4C). Detailed information regarding the GO analyses is listed in Tables 1-3.
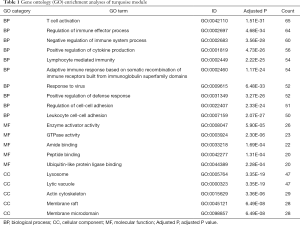
Full table
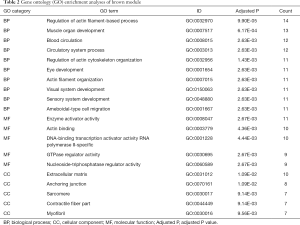
Full table

Full table
Next, we investigated the enriched pathways in the key modules. In the turquoise module, the enriched pathways were mainly associated with Herpes simplex virus 1 infection, Epstein-Barr virus infection and the phagosome. The brown module was enriched in the phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) and Ras-proximate-1 (Rap1) signaling pathways. The yellow module was enriched in the transcriptional misregulation in cancer (Figure 4D). Detailed information regarding the KEGG pathways analyses is shown in Table 4.

Full table
Identification of hub genes
In the turquoise module, integrin beta 2 (Itgb2, IC =610), tropomyosin 3, gamma (Tpm3, IC =607), Rho GTPase activating protein 30 (Arhgap30, IC =604), protein kinase C delta type (Prkcd, IC =596), and CD300C molecule 2 (CD300c2/AF251705, IC =596) were identified as hub genes (Figure 5A). In the brown module, hemecintin1 (Hmcn1, IC =186), pyruvate dehydrogenase phosphatase catalytic subunit 1 (Pdp1, IC =159), ATP-binding cassette, sub-family A (ABC1) member 8b (Abca8b, IC =158), Rab GTPase-activating protein 1-like (Rabgap1l, IC =157), and membrane-associated guanylate kinase WW and PDZ domain-containing protein 3 (Magi3, IC =157) were identified as hub genes (Figure 5B). In the yellow module, solute carrier family 9 (sodium hydrogen exchanger) member 3 regulator 2 (Slc9a3r2, IC =193), zinc finger and Broad-Complex, Tramtrack and Bric a brac (BTB) domain containing 16 (Zbtb16, IC =180), G protein-coupled receptor 56 (Gpr56, IC =179), proline and serine rich 2 (Proser2, IC =176), and midnolin (Midn, IC =175) were identified as hub genes (Figure 5C).
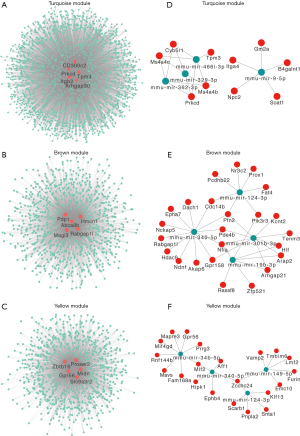
Prediction of potential miRNA-target regulatory networks
Here, we used miRNet, a tool that integrates data from 11 different miRNA databases, to predict regulatory miRNAs of the key modules. In the turquoise module, 141 miRNAs were predicted, of which the top 4 (ranked by degrees) were mmu-mir-9-5p (degree =5), mmu-mir-466i-3p (degree =5) mmu-mir-362-3p (degree =4), and mmu-mir-329-3p (degree =4) (Figure 5D). In the brown module, 180 miRNAs were predicted, of which the top 4 miRNAs were mmu-mir-340- 5p (degree =12), mmu-mir-301b- 3p (degree =9), mmu-mir-124-3p (degree =7), and mmu-mir-19b-3p (degree =7) (Figure 5E). In the yellow module, 122 miRNAs were predicted, of which the top 4 were mmu-mir-34b-5p (degree =8), mmu-mir-149-5p (degree =6), mmu-mir-124-3p (degree =5), and mmu-mir-340-5p (degree =5) (Figure 5F). Surprisingly, mmu-mir-340-5p and mmu-mir-124-3p were predicted in both the brown and yellow modules, suggesting that these 2 miRNAs may be important in chronic viral myocarditis.
Discussion
It has been proposed that high-performance “omics” and bioinformatics approaches can provide unbiased information on potential genes that are important in virus pathogenesis or host defense (1). Here, we used WGCNA to identify stage-specific gene co-expression modules and uncover novel hub genes. Exploring the occurrence and development of viral myocarditis using a biological network approach may be helpful in identifying key targets. To the best of our knowledge, this is the first gene co-expression network study to analyse viral myocarditis.
In the acute infection stage, gene expression was significantly increased in the turquoise module. Moreover, the most enriched GO terms for BP were associated with antiviral and immune-inflammatory activation. It has been reported that the virus replicates significantly after infection, causing early virus-induced cardiomyocyte injury. Neutralizing antibodies appear around 1 week post-infection and play critical roles in limiting further viral replication in the heart. After the release of virus progeny into the interstitium, natural killer (NK) cells and macrophages migrate to the sites of injury and release a large amount of proinflammatory cytokines (1,7). Consistent with our analysis, this process primarily involves anti-viral and immune responses.
Based on the intramodular connectivity, we identified five hub genes (Itgb2, Tpm3, Arhgap30, Prkcd, CD300c2) in the turquoise module. Itgb2, also known as CD18, is a member of the integrin family that is critical for mounting effective immune responses (26,27). CD11/CD18 heterodimers (Mac-1) play critical roles in leukocyte trafficking, immune synapse formation, and co–stimulation (28). Itgb2 mutations causes leukocyte extravasation deficiency that impairs their migration to sites of inflammation (29). Previous studies show that Itgb2 may be involved in the regulation of B. burgdorferi-induced carditis and autoimmune carditis by recruiting macrophages (30,31). However, the role of Itgb2 in viral myocarditis needs to be further verified. Tpm3 modulates myosin-actin interaction for cardiac contraction (32). Previous studies have shown that CVB3 infection triggers a strong inflammatory response and production of autoantibodies to cardiac antigens. Significant increases in anti-myosin heavy chain, anti-actin, and anti-tropomyosin antibodies were seen in infected mice as early as day seven post-infection. Cardiac myosin-specific autoantibodies may have an immunopathogenic role in viral myocarditis (33,34). Arhgap30 is a RhoA- and Rac1-specific Rho GAP that modulates the cytoskeleton organisation and cell adhesion (35). It has been reported that it promotes p53 acetylation (36). Furthermore, an increase of acetylated p53 is related to apoptosis in CVB3-infected cardiomyocytes (37). Therefore, the increased expression of Arhgap30 may promote apoptosis in the early stage of viral myositis. Prkcd is a critical regulator of the inflammatory response in cancer, diabetes, ischaemic heart disease, sepsis, and neurodegenerative diseases (38), and its activation is associated with myocardial ischaemia-reperfusion injury (39). Furthermore, it has been previously shown that Prkcd participates in myocardial fibrosis in autoimmune myocarditis (40,41). CD300c2 (also MAIR-II, LMIR2, or CLM-4) is an Ig-like receptor expressed on macrophages, monocytes, and a subset of B cells (42,43). It has been reported that CD300c2 deficiency in a mouse sepsis model could reduce monocyte migration and suppress proinflammatory cytokine production (44,45). Furthermore, CD300c2 contributes to bleomycin-induced inflammatory responses (43). Importantly, the predicted hub genes indicate that that virus-mediated immune responses and inflammatory damage may play a major role during acute viral myocarditis.
In the chronic disease stage, gene expression was significantly increased in the brown module, while it was significantly decreased in the yellow module. In the brown module, the most enriched GO terms for BP were mainly associated with the cytoskeleton and the regulation of actin. In the yellow module, the most enriched GO terms of BP were mainly associated with phosphorylation, cellular catabolic process, vascular regulation and autophagy. Thus, the main BP changed significantly in the chronic disease stage, compared with the acute disease stage. Previous studies have shown that the chronic stage of viral myocarditis mainly includes myocardial fibrosis and cardiac remodelling, but the specific mechanism is unclear (1,2). Therefore, our analysis may offer a new direction in the exploration of viral myocarditis.
In the brown module, we identified five hub genes (Hmcn1, Pdp1, Abca8b, Rabgap1l, Magi3). The protein encoded by Hmcn1, also known as Fibulin-6, increases significantly in myocardial infarction and participates in the regulation of cardiac remodelling after myocardial infarction (46,47). Thus, Fibulin-6 might regulate fibrosis and cardiac remodelling in myocarditis. Pdp1 resides in the mitochondrial matrix and is responsible for dephosphorylation and reactivation of pyruvate dehydrogenase, thereby modulating the utilisation of carbohydrates in mammals (48). In heart failure, nicotinamide adenine dinucleotide (NADH)-dependent mitochondrial complex I activity and oxidative phosphorylation have been found to be defective, while pyruvate dehydrogenase complex activity is significantly enhanced (49). Therefore, Pdp1 may play an important role in the regulation of cardiac energy metabolism in chronic viral myocarditis. Abca8b is a membrane-associated protein belonging to the ATP-binding cassette transporters superfamily. Abca8b acts as a cholesterol transporter and may, therefore, be involved in atherosclerosis (50). Inhibition of miR-92a-3p has been is reported to regulate cardiomyocyte metabolic switching through Abca8b, thereby improving function and recovery of endothelial cells after acute myocardial infarction (51). Rabgap1l can modulate the migration of fibroblasts (52). Magi3 is associated with various diseases, including inflammatory bowel disease, thyroid disease, breast cancer, and colon cancer (53-56). Magi3 interacts with the Beta-1 and Beta-2 adrenergic receptors, selectively regulating the signal transduction of certain receptors (57,58).
In the yellow module, Slc9a3r2, Zbtb16, Gpr56, Proser2, and Midn were identified as hub genes. The protein encoded by Slc9a3r2, also known as NHERF2, plays an important role in cell signal transduction, ion transport, membrane targeting and transport of receptors (59,60). The protein encoded by Zbtb16, also known as PLZF, is a transcription factor containing 9 zinc fingers in the carboxyl terminal area. In spontaneously hypertensive rats, reduced expression of PLZF is associated with an improvement of cardiac hypertrophy and fibrosis (61). PLZF is an important angiotensin II receptor 2 binding protein and PLZF knockout mice are resistant to angiotensin-induced cardiac hypertrophy and fibrosis (62). Furthermore, PLZF may modulate congenital heart disease (63). Gpr56 is involved in a variety of BPs, including tumorigenesis, nervous system development, regulation of islet beta cells and muscle regulation of mechanical overload (64-69). In addition, GPR56 is reported to promote cardiomyocyte hypertrophy induced by angiotensin II, leading to cardiac hypertrophy (70). Proser2 is rich in serine and proline residues that maybe serve as phosphorylation sites. Although, Proser2 has been proposed as a breast cancer biomarker (71), its functions remain unclear. Midn is involved in the regulation of genes related to neurogenesis during mouse development (72), and promotes the expression of Parkin E3 ubiquitin ligase, whose down-modulation is associated with Parkinson's disease (73). Midn has been reported to interact with glucokinase through an N-terminal ubiquitin-like domain, resulting in a significant reduction in glucokinase activity and glucose-induced insulin secretion (74). In conclusion, the hub genes in the brown and yellow modules indicate that myocardial fibrosis, abnormal energy metabolism, cellular function, and cytoskeleton changes may be involved in chronic viral myocarditis.
Previous studies have shown that miRNAs also play key roles in viral myocarditis (75-79). Therefore, we predicted potential miRNA-targeted regulatory networks in the three key modules. Of these, we found that mmu-mir-340-5p and mmu-mir-124-3p may play an important role in chronic myocarditis.
In our study, the hub genes were identified according to intramodular connectivity. Generally speaking, genes with higher intramodular connectivity are often at the core position in the co-expression module, and play an important role in the development of the disease. However, some “hub genes” may show high intramodular connectivity due to the regulation of their upstream hub genes. These regulated “hub genes” may be significantly associated with the disease, but they are not the primary drivers of disease. Therefore, our results need further verification. Furthermore, rare interactions might have been obscured due to the small sample size available for analysis.
Conclusions
Our results reveal the main biological changes in different stages of viral myocarditis, and predict the important hub genes and miRNAs in both the acute and chronic disease stages. These findings provide novel insights into the pathogenesis of viral myocarditis, which might be helpful for developing new therapeutic targets in the future.
Acknowledgments
Funding: This work was supported by grants from the National Natural Science Foundation of China (Project No. 81974025, Project No. 81570246); Natural Science Foundation of Zhejiang Province (Project No. LY19H020007); Medical and Health Science Program of Zhejiang Province (Project No. 2019323962, Project No. 2020381788); Natural Key Research and Development Project of Zhejiang Province (Project No. 2018C03015).
Footnote
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-3337
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3337). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study did not involve experiments in humans or animals. The raw dataset is available from the GEO database (http://www.ncbi.nlm.nih.gov/geo/; GSE35182).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Esfandiarei M, McManus BM. Molecular biology and pathogenesis of viral myocarditis. Annu Rev Pathol 2008;3:127-55. [Crossref] [PubMed]
- Fung G, Luo H, Qiu Y, et al. Myocarditis. Circ Res 2016;118:496-514. [Crossref] [PubMed]
- Cooper LT Jr. Myocarditis. N Engl J Med 2009;360:1526-38. [Crossref] [PubMed]
- Kindermann I, Barth C, Mahfoud F, et al. Update on myocarditis. J Am Coll Cardiol 2012;59:779-92. [Crossref] [PubMed]
- Heymans S, Eriksson U, Lehtonen J, et al. The Quest for New Approaches in Myocarditis and Inflammatory Cardiomyopathy. J Am Coll Cardiol 2016;68:2348-64. [Crossref] [PubMed]
- Yajima T, Knowlton KU. Viral myocarditis: from the perspective of the virus. Circulation 2009;119:2615-24. [Crossref] [PubMed]
- Liu PP, Mason JW. Advances in the understanding of myocarditis. Circulation 2001;104:1076-82. [Crossref] [PubMed]
- Barabási AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet 2004;5:101-13. [Crossref] [PubMed]
- Furlong LI. Human diseases through the lens of network biology. Trends Genet 2013;29:150-9. [Crossref] [PubMed]
- Chen J, Yu L, Zhang S, et al. Network Analysis-Based Approach for Exploring the Potential Diagnostic Biomarkers of Acute Myocardial Infarction. Front Physiol 2016;7:615. [Crossref] [PubMed]
- Yin L, Cai Z, Zhu B, et al. Identification of Key Pathways and Genes in the Dynamic Progression of HCC Based on WGCNA. Genes (Basel) 2018;9:92. [Crossref] [PubMed]
- Liang JW, Fang ZY, Huang Y, et al. Application of Weighted Gene Co-Expression Network Analysis to Explore the Key Genes in Alzheimer's Disease. J Alzheimers Dis 2018;65:1353-64. [Crossref] [PubMed]
- Doering TA, Crawford A, Angelosanto JM, et al. Network analysis reveals centrally connected genes and pathways involved in CD8+ T cell exhaustion versus memory. Immunity 2012;37:1130-44. [Crossref] [PubMed]
- Liu Q, Jiang C, Xu J, et al. Genome-Wide Temporal Profiling of Transcriptome and Open Chromatin of Early Cardiomyocyte Differentiation Derived From hiPSCs and hESCs. Circ Res 2017;121:376-91. [Crossref] [PubMed]
- Meng Q, Wang K, Brunetti T, et al. The DGCR5 long noncoding RNA may regulate expression of several schizophrenia-related genes. Sci Transl Med 2018;10:eaat6912. [Crossref] [PubMed]
- Coronado MJ, Brandt JE, Kim E, et al. Testosterone and interleukin-1beta increase cardiac remodeling during coxsackievirus B3 myocarditis via serpin A 3n. Am J Physiol Heart Circ Physiol 2012;302:H1726-36. [Crossref] [PubMed]
- Gautier L, Cope L, Bolstad BM, et al. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20:307-15. [Crossref] [PubMed]
- Shen Z, Lu J, Wei J, et al. Investigation of the underlying hub genes and mechanisms of reperfusion injury in patients undergoing coronary artery bypass graft surgery by integrated bioinformatic analyses. Ann Transl Med 2019;7:664. [Crossref] [PubMed]
- Pfohl SR, Kim RB, Coan GS, et al. Unraveling the Complexity of Amyotrophic Lateral Sclerosis Survival Prediction. Front Neuroinform 2018;12:36. [Crossref] [PubMed]
- Troyanskaya O, Cantor M, Sherlock G, et al. Missing value estimation methods for DNA microarrays. Bioinformatics 2001;17:520-5. [Crossref] [PubMed]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 2008;9:559. [Crossref] [PubMed]
- Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol 2005;4:Article17.
- Wang T, Zheng X, Li R, et al. Integrated bioinformatic analysis reveals YWHAB as a novel diagnostic biomarker for idiopathic pulmonary arterial hypertension. J Cell Physiol 2019;234:6449-62. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 2012;16:284-7. [Crossref] [PubMed]
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for inte-grated models of biomolecular interaction networks. Genome Res 2003;13:2498-504. [Crossref] [PubMed]
- Pribila JT, Quale AC, Mueller KL, et al. Integrins and T cell-mediated immunity. Annu Rev Immunol 2004;22:157-80. [Crossref] [PubMed]
- Luo BH, Springer TA. Integrin structures and conformational signaling. Curr Opin Cell Biol 2006;18:579-86. [Crossref] [PubMed]
- Crozat K, Eidenschenk C, Jaeger BN, et al. Impact of beta2 integrin deficiency on mouse natural killer cell development and function. Blood 2011;117:2874-82. [Crossref] [PubMed]
- Leon-Rico D, Aldea M, Sanchez R, et al. Brief report: reduced expression of CD18 leads to the in vivo expansion of hematopoietic stem cells in mouse bone marrow. Stem Cells 2014;32:2794-8. [Crossref] [PubMed]
- Guerau-de-Arellano M, Alroy J, Huber BT. Beta2 integrins control the severity of murine Lyme carditis. Infect Immun 2005;73:3242-50. [Crossref] [PubMed]
- Haasken S, Auger JL, Binstadt BA. Absence of beta2 integrins impairs regulatory T cells and exacerbates CD4+ T cell-dependent autoimmune carditis. J Immunol 2011;187:2702-10. [Crossref] [PubMed]
- Vanburen P, Palmer BM. Cooperative activation of the cardiac myofilament: the pivotal role of tropomyosin. Circulation 2010;121:351-3. [Crossref] [PubMed]
- O'Donoghue HL, Lawson CM, Reed WD. Autoantibodies to cardiac myosin in mouse cytomegalovirus myocarditis. Immunology 1990;71:20-8. [PubMed]
- Latif N, Zhang H, Archard LC, et al. Characterization of anti-heart antibodies in mice after infection with coxsackie B3 virus. Clin Immunol 1999;91:90-8. [Crossref] [PubMed]
- Naji L, Pacholsky D, Aspenstrom P. ARHGAP30 is a Wrch-1-interacting protein involved in actin dynamics and cell adhesion. Biochem Biophys Res Commun 2011;409:96-102. [Crossref] [PubMed]
- Wang J, Qian J, Hu Y, et al. ArhGAP30 promotes p53 acetylation and function in colorectal cancer. Nat Commun 2014;5:4735. [Crossref] [PubMed]
- Jiang D, Li M, Yu Y, et al. microRNA-34a aggravates coxsackievirus B3-induced apoptosis of cardiomyocytes through the SIRT1-p53 pathway. J Med Virol 2019;91:1643-51. [Crossref] [PubMed]
- Yang Q, Langston JC, Tang Y, et al. The Role of Tyrosine Phosphorylation of Protein Kinase C Delta in Infection and Inflammation. Int J Mol Sci 2019;20:1498. [Crossref] [PubMed]
- Inagaki K, Chen L, Ikeno F, et al. Inhibition of delta-protein kinase C protects against reperfusion injury of the ischemic heart in vivo. Circulation 2003;108:2304-7. [Crossref] [PubMed]
- Su Z, Yin J, Wang T, et al. Up-regulated HMGB1 in EAM directly led to collagen deposition by a PKCbeta/Erk1/2-dependent pathway: cardiac fibroblast/myofibroblast might be another source of HMGB1. J Cell Mol Med 2014;18:1740-51. [Crossref] [PubMed]
- Liu Y, Zhu H, Su Z, et al. IL-17 contributes to cardiac fibrosis following experimental autoimmune myocarditis by a PKCbeta/Erk1/2/NF-kappaB-dependent signaling pathway. Int Immunol 2012;24:605-12. [Crossref] [PubMed]
- Yotsumoto K, Okoshi Y, Shibuya K, et al. Paired activating and inhibitory immuno-globulin-like receptors, MAIR-I and MAIR-II, regulate mast cell and macrophage activation. J Exp Med 2003;198:223-33. [Crossref] [PubMed]
- Nakazawa Y, Ohtsuka S, Nakahashi-Oda C, et al. Cutting Edge: Involvement of the Immunoreceptor CD300c2 on Alveolar Macrophages in Bleomycin-Induced Lung Fibro-sis. J Immunol 2019;203:3107-11. [Crossref] [PubMed]
- Nakahashi C, Tahara-Hanaoka S, Totsuka N, et al. Dual assemblies of an activating immune receptor, MAIR-II, with ITAM-bearing adapters DAP12 and FcRgamma chain on peritoneal macrophages. J Immunol 2007;178:765-70. [Crossref] [PubMed]
- Totsuka N, Kim YG, Kanemaru K, et al. Toll-like receptor 4 and MAIR-II/CLM-4/LMIR2 immunoreceptor regulate VLA-4-mediated inflammatory monocyte migration. Nat Commun 2014;5:4710. [Crossref] [PubMed]
- Chowdhury A, Herzog C, Hasselbach L, et al. Expression of fibulin-6 in failing hearts and its role for cardiac fibroblast migration. Cardiovasc Res 2014;103:509-20. [Crossref] [PubMed]
- Chowdhury A, Hasselbach L, Echtermeyer F, et al. Fibulin-6 regulates pro-fibrotic TGF-beta responses in neonatal mouse ventricular cardiac fibroblasts. Sci Rep 2017;7:42725. [Crossref] [PubMed]
- Huang B, Gudi R, Wu P, et al. Isoenzymes of pyruvate dehydrogenase phosphatase. DNA-derived amino acid sequences, expression, and regulation. J Biol Chem 1998;273:17680-8. [Crossref] [PubMed]
- Sheeran FL, Angerosa J, Liaw NY, et al. Adaptations in Protein Expression and Regulated Activity of Pyruvate Dehydrogenase Multienzyme Complex in Human Systolic Heart Failure. Oxid Med Cell Longev 2019;2019:4532592. [Crossref] [PubMed]
- Trigueros-Motos L, van Capelleveen JC, Torta F, et al. ABCA8 Regulates Cholesterol Efflux and High-Density Lipoprotein Cholesterol Levels. Arterioscler Thromb Vasc Biol 2017;37:2147-55. [Crossref] [PubMed]
- Rogg EM, Abplanalp WT, Bischof C, et al. Analysis of Cell Type-Specific Effects of MicroRNA-92a Provides Novel Insights Into Target Regulation and Mechanism of Action. Circulation 2018;138:2545-58. [Crossref] [PubMed]
- Qu F, Lorenzo DN, King SJ, et al. Ankyrin-B is a PI3P effector that promotes polarized alpha5beta1-integrin recycling via recruiting RabGAP1L to early endosomes. Elife 2016;5:e20417. [Crossref] [PubMed]
- Banerji S, Cibulskis K, Rangel-Escareno C, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 2012;486:405-9. [Crossref] [PubMed]
- Norén E, Almer S, Soderman J. Genetic variation and expression levels of tight junction genes identifies association between MAGI3 and inflammatory bowel disease. BMC Gastroenterol 2017;17:68. [Crossref] [PubMed]
- Medici M, Porcu E, Pistis G, et al. Identification of novel genetic Loci associated with thyroid peroxidase antibodies and clinical thyroid disease. PLoS Genet 2014;10:e1004123. [Crossref] [PubMed]
- Lee SJ, Ritter SL, Zhang H, et al. MAGI-3 competes with NHERF-2 to negatively regulate LPA2 receptor signaling in colon cancer cells. Gastroenterology 2011;140:924-34. [Crossref] [PubMed]
- Yang X, Zheng J, Xiong Y, et al. Beta-2 adrenergic receptor mediated ERK activation is regulated by interaction with MAGI-3. FEBS Lett 2010;584:2207-12. [Crossref] [PubMed]
- He J, Bellini M, Inuzuka H, et al. Proteomic analysis of beta1-adrenergic receptor interactions with PDZ scaffold proteins. J Biol Chem 2006;281:2820-7. [Crossref] [PubMed]
- Shenolikar S, Weinman EJ. NHERF: targeting and trafficking membrane proteins. Am J Physiol Renal Physiol 2001;280:F389-95. [Crossref] [PubMed]
- Mederos Y, Schnitzler M, Gudermann T, Storch U. Emerging Roles of Diacylglycerol-Sensitive TRPC4/5 Channels. Cells 2018;7:218. [Crossref] [PubMed]
- Liška F, Landa V, Zidek V, et al. Downregulation of Plzf Gene Ameliorates Metabolic and Cardiac Traits in the Spontaneously Hypertensive Rat. Hypertension 2017;69:1084-91. [Crossref] [PubMed]
- Wang N, Frank GD, Ding R, et al. Promyelocytic leukemia zinc finger protein acti-vates GATA4 transcription and mediates cardiac hypertrophic signaling from angiotensin II receptor 2. PLoS One 2012;7:e35632. [Crossref] [PubMed]
- McKean DM, Homsy J, Wakimoto H, et al. Loss of RNA expression and al-lele-specific expression associated with congenital heart disease. Nat Commun 2016;7:12824. [Crossref] [PubMed]
- Piao X, Hill RS, Bodell A, et al. G protein-coupled receptor-dependent development of human frontal cortex. Science 2004;303:2033-6. [Crossref] [PubMed]
- Koirala S, Jin Z, Piao X, et al. GPR56-regulated granule cell adhesion is essential for rostral cerebellar development. J Neurosci 2009;29:7439-49. [Crossref] [PubMed]
- Yang L, Friedland S, Corson N, et al. GPR56 inhibits melanoma growth by internalizing and degrading its ligand TG2. Cancer Res 2014;74:1022-31. [Crossref] [PubMed]
- White JP, Wrann CD, Rao RR, et al. G protein-coupled receptor 56 regulates mechanical overload-induced muscle hypertrophy. Proc Natl Acad Sci U S A 2014;111:15756-61. [Crossref] [PubMed]
- Daria D, Kirsten N, Muranyi A, et al. GPR56 contributes to the development of acute myeloid leukemia in mice. Leukemia 2016;30:1734-41. [Crossref] [PubMed]
- Olaniru OE, Pingitore A, Giera S, et al. The adhesion receptor GPR56 is activated by extracellular matrix collagen III to improve beta-cell function. Cell Mol Life Sci 2018;75:4007-19. [Crossref] [PubMed]
- Zhang Y, Si Y, Ma N, et al. The RNA-binding protein PCBP2 inhibits Ang II-induced hypertrophy of cardiomyocytes though promoting GPR56 mRNA degeneration. Biochem Biophys Res Commun 2015;464:679-84. [Crossref] [PubMed]
- Sui Y, Ju C, Shao B. A lymph node metastasis-related protein-coding genes combining with long noncoding RNA signature for breast cancer survival prediction. J Cell Physiol 2019;234:20036-45. [Crossref] [PubMed]
- Tsukahara M, Suemori H, Noguchi S, et al. Novel nucleolar protein, midnolin, is ex-pressed in the mesencephalon during mouse development. Gene 2000;254:45-55. [Crossref] [PubMed]
- Obara Y, Imai T, Sato H, et al. Midnolin is a novel regulator of parkin expression and is associated with Parkinson's Disease. Sci Rep 2017;7:5885. [Crossref] [PubMed]
- Hofmeister-Brix A, Kollmann K, Langer S, et al. Identification of the ubiquitin-like domain of midnolin as a new glucokinase interaction partner. J Biol Chem 2013;288:35824-39. [Crossref] [PubMed]
- Zhang X, Gao X, Hu J, et al. ADAR1p150 Forms a Complex with Dicer to Promote miRNA-222 Activity and Regulate PTEN Expression in CVB3-Induced Viral Myocarditis. Int J Mol Sci 2019;20:407. [Crossref]
- Besler C, Urban D, Watzka S, et al. Endomyocardial miR-133a levels correlate with myocardial inflammation, improved left ventricular function, and clinical outcome in patients with inflammatory cardiomyopathy. Eur J Heart Fail 2016;18:1442-51. [Crossref] [PubMed]
- Ho BC, Yu SL, Chen JJ, et al. Enterovirus-induced miR-141 contributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Mi-crobe 2011;9:58-69. [Crossref] [PubMed]
- Hemida MG, Ye X, Zhang HM, et al. MicroRNA-203 enhances coxsackievirus B3 replication through targeting zinc finger protein-148. Cell Mol Life Sci 2013;70:277-91. [Crossref] [PubMed]
- Corsten MF, Papageorgiou A, Verhesen W, et al. MicroRNA profiling identifies mi-croRNA-155 as an adverse mediator of cardiac injury and dysfunction during acute viral myocarditis. Circ Res 2012;111:415-25. [Crossref] [PubMed]


