
RSPO3 is a marker candidate for predicting tumor aggressiveness in ovarian cancer
Introduction
Ovarian cancer is the third most common malignant tumor of the female reproductive system, and the leading cause of cancer-related death among all gynecologic malignancies worldwide. In 2019, it was estimated that 22,530 new cases were diagnosed and 13,980 women died of the disease in the United States (1). The corresponding figures in China from 2015 were 52,100 and 22,500, respectively (2). The histological type and clinical features of ovarian cancer are quite heterogeneous. Despite the great efforts made to promote early detection of ovarian cancer, only about 30% of patients can be diagnosed at early stages. For those patients with advanced diseases, such as International Federation of Gynecology and Obstetrics (FIGO) stage III and IV, the standard of care includes cytoreductive surgery and platinum-based chemotherapy, which are associated with significant complications and toxicity. Yet, the prognosis of these patients remains poor, with an estimated five-year survival rate of 40% (3). The high potential for peritoneal spreading and lymphatic metastasis is a major cause of treatment failure, and contributes to poor prognosis. However, there is a lack of effective biomarkers for early diagnosis, and the biomolecular mechanisms of aggressive tumor behavior are not well understood.
R-spondin 3 (RSPO3) is a secreted protein belonging to the RSPO family (RSPO1-4), which shares the common structure of a thrombospondin type 1 domain and an N-terminal cysteine-rich region (4,5). The RSPO family is expressed in human normal placenta, lung, and muscle tissues (6,7). This family participates in the regulation of cell proliferation and differentiation by activating and cooperating with the Wnt/β-catenin signaling pathway, and plays an important role in the growth of bones, muscles, blood vessels, and other tissues, as well as the formation of limbs and gonadal glands. Recent studies have shown that the RSPO family, especially RSPO3, is involved in the development of several human cancers. Theodorou et al. first identified RSPO3 as an oncogene based on the fact that transfected RSPO3 could induce carcinogenesis in a p53-deficient mammary epithelial cell line (8). Other authors reported that RSPO3 gene mutation or abnormal expression exists in various tumors, including colorectal cancer, lung adenocarcinoma, bladder cancer, and prostate cancer, which may affect tumor proliferation, invasion, metastasis, and stem cell characteristics; RSPO3 is also related to clinical stage and prognosis in cancer patients (9-12). Moreover, preliminary studies have demonstrated the therapeutic effect of targeting RSPO3-related signaling to treat colorectal cancer (13-15). Taken together, these findings suggest a tumor-promoting role and a potential therapeutic target for RSPO3 in certain types of human cancers. However, RSPO3 expression and its biological function in ovarian cancer have not been previously elucidated.
In this study, we investigated the expression and functional role of RSPO3 in ovarian cancer. Our findings suggested that RSPO3 is overexpressed in ovarian cancer, and is correlated with advanced stage and poor prognosis. RSPO3 could promote cell growth as well as aggressiveness in ovarian cancer cells. Since the activation of the common Wnt/β-catenin signaling pathway was not observed, RSPO3 is more likely to promote ovarian cancer invasiveness through activation of the PI3K/AKT pathway and modulation of the epithelial-mesenchymal transition (EMT) process. We present this study in accordance with the Materials Design Analysis Reporting (MDAR) reporting checklist (available at http://dx.doi.org/10.21037/atm-20-3731).
Methods
Patient specimens and clinical data
A total of 179 patients with high grade ovarian serous cancer (HGOSC) who received comprehensive staging or cytoreductive surgery in Sun Yat-sen University Cancer Center (SYSUCC) between 2008 and 2016 were enrolled in this study. Clinical specimens and follow-up data were obtained for all patients. All patients were female, with a mean age of 49.90 years. The fresh specimens were collected within 30 min after surgical removal and stored at −80 °C [for quantitative real-time polymerase chain reaction (qRT-PCR)] or liquid nitrogen [for western blotting (WB)] in SYSUCC’s cancer resource bank. The formalin-fixed and paraffin-embedded specimens were made into tissue arrays and used for immunohistochemistry (IHC) staining. Clinical and follow-up data were extracted from medical records. The clinical stages of these patients were determined under the 2013 FIGO staging system for ovarian, fallopian tube, and primary peritoneal cancer. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional ethical committee of SYSUCC (No.: GZR2020-075) and informed consent was obtained from all involved patients.
Cell lines and cell culture
Ovarian cancer cell lines SKOV3 (ATCC, Cat.# HTB-77, RRID:CVCL_0532, USA) and OVCAR3 (ATCC, Cat.# HTB-161, RRID:CVCL_0465, USA) were maintained at 37 °C in an incubator with 5% CO2 in their respective mediums: Dulbecco’s Modified Eagle’s Medium (DMEM, Gibco, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA) for SKOV3 and Roswell Park Memorial Medium 1640 (RPMI 1640, Gibco, USA) supplemented with 20% FBS for OVCAR3. The culture medium was replaced daily and cells were passaged when they reached approximately 80% confluency to maintain good growth characteristics.
Plasmids, lentiviral infection, and transfection
Human RSPO3 cDNA was amplified with PCR and ligated to pLenti6/V5 lentiviral vector (Invitrogen, USA). The ligation product was transferred into the prepared TOP10 E. coli competent cells (TransGen Biotech, China) and selected through Luria-Bertani (LB) solid medium containing chloramphenicol antibiotic. The amplified and purified plasmids were packaged with lentiviral particles, and then infected with SKOV3 and OVCAR3 cells. Three different siRNA segments for RSPO3 were synthesized (GenePharma, China) and transiently transfected into cells using Lipofectamine 2000 (Invitrogen, USA), two of which successfully knocked down RSPO3 expression in SKOV3 and OVCAR3 cells.
RNA extraction and qRT-PCR
Total RNA from tissues or cells was extracted using Trizol (Invitrogen, USA) and reverse transcription PCR was performed using HiScript Reverse Transcriptase (Vazyme, China) as per the manufacturer’s instructions. The mRNA expression was quantified using Green qPCR SuperMix kit (TransStart, China) on the StepOnePlus Real-time PCR system. A three-step PCR reaction was used (94 °C × 15 s, 55 °C × 30 s, 70 °C × 30 s, 45 cycles), and mRNA expression level was calculated using the comparative Ct method with GAPDH as an internal control. The following forward and reverse primer sequences were used: GAPDH (internal control), (F) AAGGAGT AAGACCCCTGGACCA, (R) GCAACTGTGAGCAGGGGAGATT; RSPO3, (F) TG TCAGTATTGTGCACTGTGAGGT, (R) TCGGACCCGTGTTTCAGTCC.
Western blotting
The tissues and cells were lysed with RIPA buffer plus protease inhibitors as per the manufacturer’s instructions. The extracted protein was quantified and subjected to SDS-PAGE (10%) gel for WB analysis. After transfer to PVDF membranes, blots were incubated with the following primary antibodies: anti-RSPO3 (Abcam, Cat.#ab233113, RRID:AB_2848140, USA), anti-p-Akt (Cell Signaling Technology, Cat.#4060, RRID:AB_2315049, USA), anti-t-Akt (Cell Signaling Technology, Cat.#4691, RRID:AB_915783, USA), anti-β-catenin (Cell Signaling Technology, Cat.#8480, RRID:AB_11127855, USA), anti-E-cadherin (Abcam, Cat.#ab40772, RRID:AB_731493, USA) and anti-GAPDH (Proteintech, Cat.#60004-1-Ig, RRID:AB_2107436, USA). Following washing with Tris Buffered Saline with Tween (TBST), blots were then incubated with polyclonal secondary antibody and imaged. The antibodies used are shown in Table 1.
Full table
Immunohistochemistry
Tissue arrays of HGOSC primary tumors were deparaffinized and hydrated. After citrate antigen repair and blocking, the sections were immunostained with anti-RSPO3 primary antibody (Abcam, Cat.#ab233113, RRID: AB_2848140, USA, shown in Table 1) and then incubated with secondary antibodies conjugated to peroxidase-labeled polymers (PV6000, Zsbio, China). The samples were finally stained with DAB and hematoxylin (Zsbio, China) and imaged under a photomicroscope. Pictures were captured and analyzed quantitatively using an automatic digital pathology system (KFBIO, China). The immunohistochemical staining intensity of RSPO3 was evaluated by three experienced pathologists independently, and scored as follows: 0, negative; 1, weak; 2, moderate; and 3, strong. Disagreement was solved by consensus conference. Normal fallopian tube tissues were treated with the same procedure and used as the normal control.
Immunofluorescence
Appropriate numbers of cells were planted on glass coverslips in a 12-well plate. After being washed with PBS, fixed in 4% paraformaldehyde, permeabilized with 0.3% TritonX-100, and blocked with 3% BSA, cells were incubated with β-catenin primary antibody (Cell Signaling Technology, Cat.#8480, RRID:AB_11127855, USA, shown in Table 1), followed by incubation with Donkey anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor Plus 488 (Thermo Fisher, USA). Finally, the glass coverslips were stained with DAPI and photographed under a fluorescence microscope.
Cell Counting Kit-8 (CCK-8) assay
CCK-8 assay was used to detect cell proliferation activity. Appropriate numbers of cells were seeded in a 96-well plate. After the plate was incubated for a specific length of time (1, 2, 3, or 4 days), CCK-8 solution (Yeasen, China) was added and incubated. Cell viability was determined by measuring absorbance at 450 nm using a microplate reader.
Colony formation assay
Cells were planted in a six-well plate in a complete medium. After 14 days of culturing to form colonies, the cells were fixed and stained with crystal violet. The colonies were photographed with a digital camera and counted using Image J software (NIH, USA).
Wound healing assay
A wound healing assay was applied to detect the capacity for cell migration. Cells were seeded in a six-well plate and incubated in a complete medium until 90% confluence was reached. The confluent cell monolayer was scratched using a pipette tip, and then incubated for 24 hours in serum-free medium. The scratch area (SA) was measured using Image J software (NIH, USA) and the wound healing rate was calculated using the following formula: wound healing rate = (SA before − SA after)/SA before.
Matrigel transwell assay
The Matrigel transwell assay was conducted to detect the capacity for cell invasion using a transwell chamber (Corning, USA). Cells were starved overnight and then seeded in the upper Matrigel (BD, USA) pre-coated chamber with serum-free medium. The lower chamber was filled with complete medium at a higher concentration as a chemoattractant. After incubation for 24 hours, cells that had invaded through the membrane were fixed and stained with crystal violet. The cells were imaged under a photomicroscope and counted using Image J software (NIH, USA).
Data downloading and bioinformatics analysis
To explore the potential mechanism of RSPO3 functioning, we obtained the publicly available RNA-Seq data of ovarian cancer patients from The Cancer Genome Atlas (TCGA, RRID: SCR_003193, https://cancergenome.nih.gov, study ID: ov_tcga_pan_can_atlas_2018, 379 samples) with the help of R software (R Project for Statistical Computing, RRID: SCR_001905, https://www.R-project.org). The possible biological processes associated with RSPO3 were identified using the Gene Set Enrichment Analysis software (GSEA 4.0.3, RRID: SCR_003199, https://www.gsea-msigdb.org) (16,17).
Transcriptome sequencing and functional enrichment analysis
The RSPO3 overexpression OVCAR3 cell group and control group were sent to undergo standard transcriptome next-generation sequencing at the HaploX Genomics Center (HGC, Shenzhen, China). Each group had three independent samples. RNA samples were quantified using a Qubit 3.0 Fluorometer (Life Technologies, CA, USA), and RNA integrity was assessed using a 4200 TapeStation (Agilent Technologies, CA, USA). The RNA sequencing library was constructed and sequenced using the Illumina PE150 platform. Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis, and visualization of differentially-expressed genes was implemented by the cluster Profiler R package, which is a simple-to-use tool to analyze high-throughput data obtained from transcriptomics or proteomics. Genes with adjusted P values less than 0.05 were considered significantly enriched by differentially-expressed genes. The raw RNA sequencing data were deposited to Sequenced Read Archive (SRA) under accession number PRJNA655466.
Statistical analysis
All experiments were repeated three times unless otherwise stated. Statistical analysis was carried out using SPSS version 16.0 (SPSS, RRID: SCR_002865, USA) and GraphPad Prism 5 (GraphPad Software, RRID: SCR_002798, USA). The mean differences between two groups were compared using Student’s t test, one-way ANOVA was used to compare three or more groups, and two-way ANOVA was used for data with a time variable. The relationship between RSPO3 expression and clinicopathological characteristics was analyzed using the chi-square test or Fisher’s exact test. Univariate survival analyses were plotted using the Kaplan-Meier survival method and analyzed with the log-rank test. A Cox proportional hazard model was used for multivariate survival analyses. Data are presented as mean values with standard deviation (SD). A two-sided P value <0.05 was used to define statistical significance.
Results
RSPO3 is upregulated in ovarian cancer tissues and correlated with aggressive clinicopathological characteristics
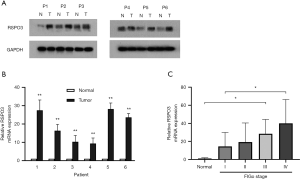
We explored the expression level of RSPO3 in six ovarian cancer tissues and their matched normal fallopian tube tissues with WB and qRT-PCR. Both results showed that RSPO3 was expressed at a significantly higher level in ovarian cancer tissues compared with normal control tissues (Figure 1A,B), which indicated RSPO3’s potential function in promoting ovarian cancer. Next, we performed qRT-PCR to analyze 40 ovarian cancer samples, and found that RSPO3 expression was positively correlated with advanced tumor stage (P<0.05 between groups, P<0.01 in summary, Figure 1C).
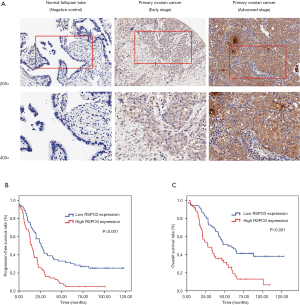
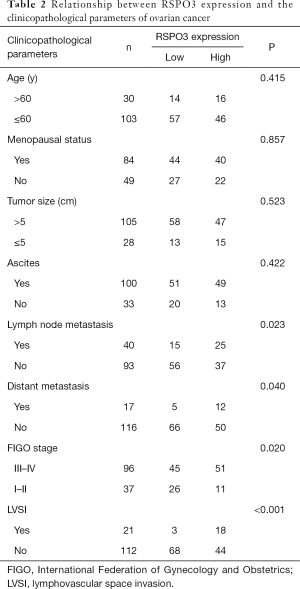
In order to further investigate the role of RSPO3 in ovarian cancer, the specimens from 133 HGOSC patients, as well as their clinical data, were collected. Through scoring IHC staining intensity, RSPO3 expression was evaluated in both tumor and corresponding normal tissues with three representatives shown (Figure 2A). A staining score of 0–2 was defined as low expression, while 3 was defined as high expression. The correlation analysis between RSPO3 expression and the clinicopathological characteristics of ovarian cancer patients showed that high RSPO3 expression was associated with lymphovascular space invasion (LVSI), lymph node metastasis, distant metastasis, and advanced tumor stage (Table 2), which revealed that RSPO3 may play an active role in the tumor invasion and progression of ovarian cancer.
Full table
RSPO3 is an independent prognostic factor for ovarian cancer patients
To determine whether RSPO3 expression had an impact on survival of ovarian cancer patients, we performed Kaplan-Meier curves for progression-free survival (PFS) and overall survival (OS), and found that high RSPO3 expression was significantly correlated with poor survival in ovarian cancer patients (P<0.001 for both PFS and OS, Figure 2B,C). In univariate and multivariate analysis using the Cox proportional hazards model, RSPO3 was found to be a strong prognostic factor for ovarian cancer patients, independent of age, menopausal status, tumor size, ascites, residual tumor, FIGO stage, or LVSI (HR =1.628, 95% CI: 1.092–2.427, P=0.017, Table 3).
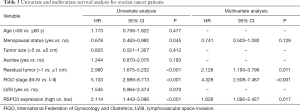
Full table
RSPO3 increases cell proliferation and colony formation of ovarian cancer in vitro
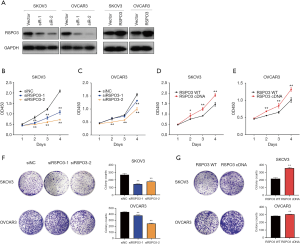
To investigate the biological function of RSPO3 in ovarian cancer, we successfully constructed the RSPO3-knockdown and RSPO3-overexpression cell model in SKOV3 and OVCAR3 cell lines (Figure 3A). CCK-8 and colony formation assays were conducted in vitro to determine the proliferation and clonogenic abilities of transfected ovarian cancer cells. The results showed that knockdown or overexpression of RSPO3 significantly decreased or increased the proliferation rate (Figure 3B,C,D,E) and colony numbers (Figure 3F,G), respectively, in both SKOV3 and OVCAR3 cells, which indicated that RSPO3 could play an important role in the formation and development of ovarian cancer.
RSPO3 promotes cell migration and invasion of ovarian cancer in vitro
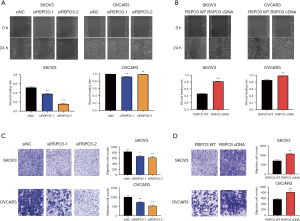
The results from our clinical specimens showed that high RSPO3 expression was correlated with LVSI, lymph node metastasis, distant metastasis, and advanced tumor stage in ovarian cancer (Figure 1C and Table 2). To further investigate whether RSPO3 contributed to the metastasis process, a wound healing assay and transwell experiment were carried out in transfected ovarian cancer cells to explore the migration and invasion abilities, respectively. The results illustrated that RSPO3 knockdown remarkably suppressed cancer cell migration and invasion in SKOV3 and OVCAR3 cells, while RSPO3 overexpression significantly enhanced these abilities (Figure 4A,B,C,D), revealing that RSPO3 may be crucial to the aggressiveness of ovarian cancer.
PI3K/AKT pathway and EMT are involved in RSPO3 signaling
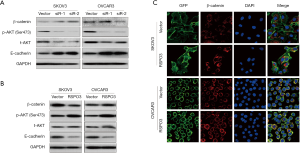
To explore the potential mechanism through which RSPO3 promotes tumor aggressiveness in ovarian cancer, we extracted the RNA-Seq data from TCGA’s ovarian cancer cohort (study ID: ov_tcga_pan_can_atlas_2018, 379 samples) and identified possible biological processes associated with RSPO3 using the GSEA method. We found that high RSPO3 expression was significantly correlated with activation of the PI3K/AKT pathway and the EMT phenomenon (Figure 5A,B), both of which have been reported to play important roles in tumor progression and metastasis (18-20). Since previous studies have stated that RSPO3 may act through Wnt/β-catenin signaling (9,11,21), we also analyzed this pathway in our study. However, the results did not show a significant connection between RSPO3 expression and Wnt/β-catenin signaling (Figure 5C), which indicated that other novel functioning pathways were involved in RSPO3’s regulation of ovarian cancer.
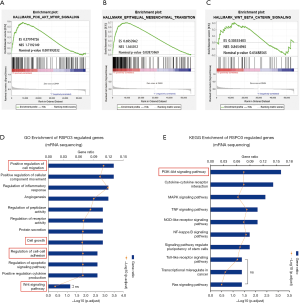
To directly observe RSPO3’s regulation process in ovarian cancer, the RSPO3 overexpression group of OVCAR3 cells and the control group were sent to undergo transcriptome next-generation sequencing. The differentially expressed genes between the groups were detected, and the pathways correlated with RSPO3 overexpression were enriched by GO and KEGG functional analysis. Overall, 113 GO items and seven KEGG items were found to be statistically significant (P<0.05 and gene counts ≥5). The top 11 GO items and top 10 KEGG items (ranked by gene ratio) are shown in Figure 5D and E, respectively. The results showed that RSPO3 had a significant influence on cell growth and migration abilities (Figure 5D), which was consistent with our previous observations in cell models. Moreover, the PI3K/AKT pathway and cell adhesion, a key biological feature involved in EMT, were shown to be significantly affected by RSPO3 overexpression (Figure 5D,E). Additionally, Wnt signaling did not appear to be involved in RSPO3 functioning (Figure 5D), a finding that was similar to the results from TCGA’s ovarian cancer cohort.
Based on the above findings, we detected several key molecules involved in the PI3K/AKT pathway and EMT in ovarian cancer cells and assessed the effect of RSPO3 on the subcellular localization of β-catenin in order to further determine the underlying mechanism of RSPO3 function. As shown in Figure 6A and B, RSPO3 overexpression greatly enhanced AKT phosphorylation (which is a key process in PI3K/AKT activation) and decreased E-cadherin expression (which is a marker of reduced cellular adhesion and EMT promotion), while RSPO3 knockdown showed the opposite effects in ovarian cancer cells. Additionally, RSPO3 expression did not induce significant changes in β-catenin nucleus transportation (which is an important aspect of Wnt/β-catenin signaling, Figure 6C). Taken together, the results indicated that RSPO3 may promote ovarian cancer aggressiveness primarily through the PI3K/AKT pathway and EMT modulation.
Discussion
Metastasis plays a crucial role in promoting ovarian cancer progression and significantly impairs patient survival. Ovarian cancer often metastasizes throughout the peritoneal cavity, and the omentum is the most common metastatic site (22). The process of ovarian cancer metastasis is usually explained by the “seed and soil hypothesis,” which states that cancer cells (seeds) are washed away passively from primary ovarian tumor via physiological ascites circulation, then adhere to and proliferate on the preferential metastatic site (soil, usually omentum) to form new lesions (23,24). In addition, metastasis via the hematogenous route was recently proposed by scholars and initially verified in a parabiosis animal model (25). However, both patterns of metastasis involve highly complex intercellular interactions and molecular regulatory networks, and the underlying mechanisms are not yet well understood. Clarifying the molecular mechanisms underlying ovarian cancer metastasis is pivotal for its prevention, diagnosis, and prognosis, as well as clinical therapy (23).
RSPO3 is a member of the RSPO family, which includes RSPO1–4 and was originally found to be an agonist of Wnt/β-catenin signaling with critical roles in normal development and stem cell function. The involvement of RSPO3 in inducing carcinogenesis after being transfected into p53-deficient mammary epithelial cells was reported (8), indicating a potential role for RSPO3 in malignant tumor behavior. Recent studies have identified recurrent gene fusions of RSPO2 and RSPO3 in a subset of colorectal cancer (CRC), which results in overexpression of tumor-cell derived RSPO2 and RSPO3 (12). The Wnt activation of RSPO3 fusion protein was obviously correlated with tumorigenesis, and tumors harboring RSPO3 fusion were found to be highly sensitive to RSPO3-blocking antibodies (13-15). OMP-131R0 (rosmantuzumab), one of the RSPO3-neutralizing antibodies, has been a clinical-stage therapeutic agent, which has shown significant effect in inhibiting the growth rate of tumors derived from CRC patients with RSPO3 fusion (15). These findings suggested a tumor-promoting role of RSPO3 and its great potential as a therapeutic target. Although the genomic rearrangement of RSPO3 seems to be specific to CRC, the aberrant expression of RSPO3 and its contribution to human cancer have been reported in other cancer types. Gong et al. found that RSPO3 is highly expressed in a subset of lung adenocarcinoma and showed a capacity for promoting tumor growth and aggressiveness in vitro and in vivo, which may account for its significant correlation with poor survival in lung cancer patients (11). Moreover, Chen et al. reported that RSPO3 upregulation in bladder cancer patients also resulted in a survival detriment (9). However, there is a study suggesting that RSPO3 has the opposite effect in prostate cancer. RSPO3 was found to be downregulated in prostate cancer and its lower expression associated with poorer survival, while loss of RSPO3 function increased invasiveness in vitro (10). These contradictory findings indicated that RSPO3 may play a dual tumor-promoting and inhibiting role in cancer overall. The complex regulation mechanism of RSPO3 in malignancy has not yet been identified, and more studies are needed to explore this valuable molecular target for human cancer. The relationship between RSPO3 and ovarian cancer has not been previously elucidated.
In this study, we demonstrated for the first time that RSPO3 is highly expressed in ovarian cancer and that its expression level is positively correlated with tumor metastasis and disease progression. We also reported a significant trend of decreased PFS and OS in ovarian cancer patients with a higher RSPO3 expression level, indicating that RSPO3 may serve as a novel prognostic factor in ovarian cancer. These findings in clinical samples were in accordance with our observations in cell models, which showed that RSPO3 could enhance the migration and invasion abilities of ovarian cancer in vitro. Thus, this preclinical study verified the prognostic value and tumor-promoting role of RSPO3 in ovarian cancer, and demonstrated that RSPO3 may be a potential therapeutic target to further improve ovarian cancer treatment.
For several decades, biological omics technologies have revolutionized cancer research. Omics technologies include comprehensive detection of genes (genomics), RNAs (transcriptomics), proteins (proteomics), metabolites (metabolomics), and quantitative features of medical imaging (radiomics) (26). Omics technologies have been widely applied in basic research and clinical practice to help accelerate the predictive, preventive, and personalized medicine (PPPM) promoted in modern medical practice (27-29). Transcriptomics is the quantitative study of whole intracellular transcripts, addressing genome function and improving discovery of differentially-expressed genes and signaling pathways correlated with tumor initiation and development. To further explore the underlying mechanisms through which RSPO3 promotes ovarian cancer metastasis, this study used the transcriptome sequencing data from TCGA’s ovarian cancer cohort as well as our own experimental cell model, and applied functional enrichment analysis to identify significant RSPO3-related pathways. We found that the PI3K/AKT pathway and EMT process were associated with RSPO3 expression, and may thus be involved in the mechanism through which RSPO3 promotes ovarian cancer aggressiveness. The PI3K/AKT pathway has been identified as the most frequently altered pathway in ovarian cancer, and its activation has found to be associated with decreased survival of ovarian cancer patients (20,22). A previous study reported that the PI3K/AKT pathway is involved with tumor EMT, which is a crucial cellular process occurring during tumor progression, through which cancer cells lose their polarity and adhesion (epithelial phenotype) while gaining migration and invasion capabilities (mesenchymal phenotype) to promote metastasis (19,30). In agreement herewith, we found that overexpression of RSPO3 significantly increased, while knockdown of RSPO3 diminished the expression level of the key molecules in PI3K/AKT signaling and EMT in ovarian cancer cells. Thus, we speculated that RSPO3 may mediate EMT via the PI3K/AKT signaling pathway to promote ovarian cancer aggressiveness. Although our study provided the preliminary evidence supporting the association between RSPO3 and the PI3K/AKT pathway as well as EMT, the regulation mechanism of RSPO3 in ovarian cancer metastasis still needs further detailed clarification.
Previously, it was reported that RSPO3 regulates through potentiating canonical Wnt/β-catenin signaling in stem cell function as well as tumor progression (6,7,12), but recent accumulating evidence has shown that RSPO3 may also act through a β-catenin-independent non-canonical Wnt signaling pathway (31-33). Intriguingly, in our experiments, Wnt-associated signaling pathways, including both the canonical (Wnt/β-catenin pathway) and the commonly defined non-canonical Wnt signaling (Wnt/PCP/JNK pathway and Wnt/Ca2+/PKC/ERK pathway), were not found to be significantly correlated with RSPO3 in ovarian cancer, and interfering with RSPO3 expression did not affect β-catenin activity in ovarian cancer cells. These observations indicate that RSPO3 may promote ovarian cancer aggressiveness through a distinct signaling pathway not related to Wnt. Thus, based on the aforementioned preliminary evidence in our study, we put forward a novel significant pathway, the PI3K/AKT pathway, that may be involved in RSPO3 regulation in ovarian cancer.
Since RSPO3 is a type of secreted protein and needs to act on cell membrane receptors, it is worthwhile to discuss the related ligand-receptor system of RSPO3 regulation, which may provide a possible explanation for the complex downstream pathways involved, as well as the contradictory roles of RSPO3 in human cancer. The type II leucine-rich repeat-containing G protein-coupled receptors (LGRs), including LGR4, LGR5, and LGR6, have been identified as the specific receptors for the RSPO family in Wnt signaling activation (34). Although LGR belongs to the G protein-coupled receptor (GPCR) superfamily, the way it participates in potentiating Wnt signaling seems to be independent of the traditional GPCR activation mechanism, given the fact that LGR is neither coupled to heterotrimeric G proteins nor to β-arrestin when stimulated by RSPOs (35). RSPOs and LGRs were shown to act through simultaneous binding to two E3 ligases, RNF43 and ZNRF3, which affected the level of Wnt-related FZD receptors on the cell membrane, thus consequently regulating Wnt signaling activity (36-38). However, some interesting findings have suggested that there is a complicated interaction between RSPOs and these associated receptors. For instance, recent studies showed that some RSPO proteins, including RSPO3, may potentiate Wnt signaling through an LGR-independent mechanism (4,39), indicating that there may still remain some undetected receptors involved with RSPO3 regulation, as well as the great potential of RSPO3 to modulate a distinct pathway other than Wnt signaling. Furthermore, some studies have pointed out that many of the downstream gene products are common molecules between the canonical and non-canonical Wnt signaling, and these molecules simultaneously have multiple interactions with other signaling pathways (21,40), and thus form a complicated regulation network and mechanistic basis for RSPO3 action. Taken together, the still unclear ligand-receptor system and the complex interactive regulation network of RSPO3 signaling could explain the complexity of RSPO3 function in human cancer and its context-specific roles in different cancer types. Future studies are needed to further explore and clarify the regulation mechanisms of the RSPO family.
This study is a preliminary exploration of the role of RSPO3 in ovarian cancer, which adds new knowledge about the molecular regulatory mechanisms involved in tumor progression. Given that RSPO3 monoclonal antibody has already been proven effective in the patient-derived tumor xenograft (PDX) models of colon cancer and acute myeloid leukemia (15,41), our study provides an initial theoretical basis for the preclinical use of this promising drug in ovarian cancer. However, a few limitations of this study must be addressed. First, tumor formation experiments in animal models are still needed to verify the tumor-promoting effect of RSPO3 in vivo. Second, the prognostic value of RSPO3 in ovarian cancer patients was based on analysis of retrospective cohorts and should be further validated in prospective studies. In addition, the regulatory mechanism of RSPO3 in ovarian cancer metastasis still needs further detailed clarification. Since RSPO3 is a secreted protein, the key molecules on the cell membrane with which RSPO3 binds to activate the downstream PI3K/AKT pathway, and the exact signaling molecules involved in the RSPO3/PI3K/AKT pathway remain unclear and warrant further research.
Conclusions
RSPO3 is upregulated in ovarian cancer, and its high expression is correlated with tumor metastasis and poor survival. Using transcriptome analysis, we provided preliminary evidence that RSPO3 can mediate EMT via the PI3K/AKT signaling pathway to promote ovarian cancer aggressiveness. However, future studies are still required to further clarify the tumor-promoting mechanism of RSPO3 in ovarian cancer. Given that targeting RSPO3 may serve as an alternative therapeutic strategy, it is worthy of further exploration in experimental animal models as well as in well-designed clinical trials. Currently, there is no ideal biomarker for ovarian cancer. The use of modern multi-omics technology may be an effective approach to discover the molecular mechanisms of carcinogenesis on multiple biological levels, and thus, this approach deserves more attention from clinicians and medical researchers.
Acknowledgments
We thank LetPub (www.letpub.com) for its linguistic assistance during the preparation of this manuscript.
Funding: This study was supported in part by the National Natural Science Foundation of China (No. 81772782).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-3731
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-3731
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-3731). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the institutional ethical committee of SYSUCC (No.: GZR2020-075) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69:7-34. [Crossref] [PubMed]
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016;66:115-32. [Crossref] [PubMed]
- Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin 2018;68:284-96. [Crossref] [PubMed]
- Park S, Cui J, Yu W, et al. Differential activities and mechanisms of the four R-spondins in potentiating Wnt/β-catenin signaling. J Biol Chem 2018;293:9759-69. [Crossref] [PubMed]
- Chen JZ, Wang S, Tang R, et al. Cloning and identification of a cDNA that encodes a novel human protein with thrombospondin type I repeat domain, hPWTSR. Mol Biol Rep 2002;29:287-92. [Crossref] [PubMed]
- Kim KA, Zhao J, Andarmani S, et al. R-Spondin proteins: a novel link to beta-catenin activation. Cell Cycle 2006;5:23-6. [Crossref] [PubMed]
- Kim KA, Wagle M, Tran K, et al. R-Spondin family members regulate the Wnt pathway by a common mechanism. Mol Biol Cell 2008;19:2588-96. [Crossref] [PubMed]
- Theodorou V, Kimm MA, Boer M, et al. MMTV insertional mutagenesis identifies genes, gene families and pathways involved in mammary cancer. Nat Genet 2007;39:759-69. [Crossref] [PubMed]
- Chen Z, Zhou L, Chen L, et al. RSPO3 promotes the aggressiveness of bladder cancer via Wnt/beta-catenin and Hedgehog signaling pathways. Carcinogenesis 2019;40:360-9. [Crossref] [PubMed]
- Mesci A, Lucien F, Huang X, et al. RSPO3 is a prognostic biomarker and mediator of invasiveness in prostate cancer. J Transl Med 2019;17:125. [Crossref] [PubMed]
- Gong X, Yi J, Carmon KS, et al. Aberrant RSPO3-LGR4 signaling in Keap1-deficient lung adenocarcinomas promotes tumor aggressiveness. Oncogene 2015;34:4692-701. [Crossref] [PubMed]
- Seshagiri S, Stawiski EW, Durinck S, et al. Recurrent R-spondin fusions in colon cancer. Nature 2012;488:660-4. [Crossref] [PubMed]
- Fischer MM, Yeung VP, Cattaruzza F, et al. RSPO3 antagonism inhibits growth and tumorigenicity in colorectal tumors harboring common Wnt pathway mutations. Sci Rep 2017;7:15270. [Crossref] [PubMed]
- Storm EE, Durinck S, de Sousa EMF, et al. Targeting PTPRK-RSPO3 colon tumours promotes differentiation and loss of stem-cell function. Nature 2016;529:97-100. [Crossref] [PubMed]
- Chartier C, Raval J, Axelrod F, et al. Therapeutic targeting of tumor-derived R-spondinattenuates β-catenin signaling and tumorigenesis in multiple cancer types. Cancer Res 2016;76:713-23. [Crossref] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 2005;102:15545-50. [Crossref] [PubMed]
- Mootha VK, Lindgren CM, Eriksson KF, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267-73. [Crossref] [PubMed]
- Xu Z, Han X, Ou D, et al. Targeting PI3K/AKT/mTOR-mediated autophagy for tumor therapy. Appl Microbiol Biotechnol 2020;104:575-87. [Crossref] [PubMed]
- Loret N, Denys H, Tummers P, et al. The role of epithelial-to-mesenchymal plasticity in ovarian cancer progression and therapy resistance. Cancers (Basel) 2019;11:E838. [Crossref] [PubMed]
- Mabuchi S, Kuroda H, Takahashi R, et al. The PI3K/AKT/mTOR pathway as a therapeutic target in ovarian cancer. Gynecol Oncol 2015;137:173-9. [Crossref] [PubMed]
- Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol 2012;13:767-79. [Crossref] [PubMed]
- Cheaib B, Auguste A, Leary A. The PI3K/Akt/mTOR pathway in ovarian cancer: therapeutic opportunities and challenges. Chin J Cancer 2015;34:4-16. [Crossref] [PubMed]
- Lengyel E. Ovarian cancer development and metastasis. Am J Pathol 2010;177:1053-64. [Crossref] [PubMed]
- Naora H, Montell DJ. Ovarian Cancer Metastasis: Integrating insights from disparate model organisms. Nat Rev Cancer 2005;5:355-66. [Crossref] [PubMed]
- Pradeep S, Kim SW, Wu SY, et al. Hematogenous metastasis of ovarian cancer: rethinking mode of spread. Cancer Cell 2014;26:77-91. [Crossref] [PubMed]
- Lu M, Zhan X. The crucial role of multiomic approach in cancer research and clinically relevant outcomes. EPMA Journal 2018;9:77-102. [Crossref] [PubMed]
- Janssens JP, Schuster K, Voss A. Preventive, predictive, and personalized medicine for effective and affordable cancer care. EPMA Journal 2018;9:113-23. [Crossref] [PubMed]
- Li N, Zhan X. Identification of clinical trait-related lncRNA and mRNA biomarkers with weighted gene co-expression network analysis as useful tool for personalized medicine in ovarian cancer. EPMA Journal 2019;10:273-90. [Crossref] [PubMed]
- Li N, Zhan X. Signaling pathway network alterations in human ovarian cancers identified with quantitative mitochondrial proteomics. EPMA Journal 2019;10:153-72. [Crossref] [PubMed]
- Wu DM, Zhang T, Liu YB, et al. The PAX6-ZEB2 axis promotes metastasis and cisplatin resistance in non-small cell lung cancer through PI3K/AKT signaling. Cell Death Dis 2019;10:349. [Crossref] [PubMed]
- Glinka A, Dolde C, Kirsch N, et al. LGR4 and LGR5 are R-spondin receptors mediating Wnt/β-catenin and Wnt/PCP signalling. EMBO Rep 2011;12:1055-61. [Crossref] [PubMed]
- Zhang M, Zhang P, Liu Y, et al. RSPO3-LGR4 regulates osteogenic differentiation of human adipose-derived stem cells via ERK/FGF signalling. Sci Rep 2017;7:42841. [Crossref] [PubMed]
- Dong X, Liao W, Zhang L, et al. RSPO2 suppresses colorectal cancer metastasis by counteracting the Wnt5a/Fzd7-driven noncanonical Wnt pathway. Cancer Lett 2017;402:153-65. [Crossref] [PubMed]
- Carmon KS, Gong X, Lin Q, et al. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci USA 2011;108:11452-7. [Crossref] [PubMed]
- de Lau W, Barker N, Low TY, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondinsignalling. Nature 2011;476:293-7. [Crossref] [PubMed]
- Zebisch M, Xu Y, Krastev C, et al. Structural and molecular basis of ZNRF3/RNF43 transmembrane ubiquitin ligase inhibition by the Wnt agonist R-spondin. Nat Commun 2013;4:2787. [Crossref] [PubMed]
- Xie Y, Zamponi R, Charlat O, et al. Interaction with both ZNRF3 and LGR4 is required for the signalling activity of R-spondin. EMBO Rep 2013;14:1120-6. [Crossref] [PubMed]
- Koo BK, Spit M, Jordens I, et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature 2012;488:665-9. [Crossref] [PubMed]
- Szenker-Ravi E, Altunoglu U, Leushacke M, et al. RSPO2 inhibition of RNF43 and ZNRF3 governs limb development independently of LGR4/5/6. Nature 2018;557:564-9. [Crossref] [PubMed]
- Mesci A, Lucien F, Huang X, et al. RSPO3 is a prognostic biomarker and mediator of invasiveness in prostate cancer. J Transl Med 2019;17:125. [Crossref] [PubMed]
- Salik B, Yi H, Hassan N, et al. Targeting RSPO3-LGR4 Signaling for Leukemia Stem Cell Eradication in Acute Myeloid Leukemia. Cancer Cell 2020;38:263-78. [Crossref] [PubMed]


