
A bioinformatics analysis on the potential role of ACE2 in cardiac impairment of patients with coronavirus disease 2019
Introduction
Since December 2019, a novel pneumonia of unknown cause has swept across the world. It was later confirmed to be an acute respiratory infectious disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The World Health Organization then designated it as coronavirus disease 2019 (COVID-19) and declared the outbreak constituted a public health emergency of international concern. By March 22, 2020, the number of confirmed cases of COVID-19 across 184 countries had surpassed 308,000, and more than 9,900 patients with COVID-19 had died.
Acute cardiac injury is a common and often fatal complication of coronavirus infection. Both acute myocarditis and acute myocardial infarction were reported in cases of severe acute respiratory syndrome (SARS) (1) and Middle East respiratory syndrome (MERS) (2). Moreover, 71.9% of patients with SARS developed tachycardia, and 40% reported that their symptoms persisted during recovery (1). More evidence came from autopsies. Specifically, the myocardial tissue of patients with SARS was significantly damaged and was infiltrated with inflammatory cells (3). As SARS-CoV-2 has high similarity to SARS-CoV and MERS-CoV (4), these findings indicated that the same cardiac injury may occur in patients with COVID-19.
Although the overall case fatality rate (CFR) of COVID-19 is relatively low, it appears to have greater cardiac impacts in severe cases compared with severe cases of SARS and MERS. In a study of 138 patients with COVID-19, 7.2% (10/138) of patients developed acute cardiac injury, and the patients who were admitted to the intensive care unit (ICU) had more severe cardiac complications than did non-ICU patients (5). Another report showed that 12% (5/41) of patients with COVID-19 were diagnosed with cardiac injury after infection (6). In a retrospective cohort study, the outcome of heart failure and acute cardiac injury was observed in 23% (44/191) and 17% (33/191) of hospitalized patients with COVID-19 respectively, but the proportion of cases with adverse cardiac events, including heart failure and acute cardiac injury, was as high as 52% (28/54) and 59% (32/54) in eventual non-survivors, respectively (7). Furthermore, the abnormally increased myocardial injury marker troponin (TNI) was found in patients with COVID-19, and increasing range was related to patients’ outcomes. Specifically, TNI multiplied at an astonishing rate in the non-survivors’ terminal phase but held steady in survivors (7). The high level of myocardial injury markers was associated with increased inflammation and decreased immunity (8). Imaging diagnosis also supported this conclusion on cardiac injury, as patients with COVID-19 showed lower epicardial adipose tissue density than normal ranges of density (9). In addition, the clinical features of patients with COVID-19 indicated that cardiac complications are common and can impact mortality, while no evidence has demonstrated that SARS-CoV-2 can directly infect the heart and cause cardiac injury.
Reports have been suggested that both SARS-CoV-2 and SARS-CoV express a similar spike glycoprotein, which can strongly bind to angiotensin-converting enzyme 2 (ACE2), the virus-specific receptor expressed on the cell surface for entry into target host cells (10). In particular, the binding affinity between SARS-CoV-2 and ACE2 has been found to be approximately 10- to 20-fold higher than that between SARS-CoV and ACE2 (11). Furthermore, the receptor ACE2 expression in human tissues suggested that SARS-CoV-2 may directly invade or damage the digestive (12) and reproductive organs (13). Thus, the ACE2-expressing cells and tissues could potentially act as targets of the SARS-CoV-2 infection.
In this study, we explored the ACE2 expression pattern in mouse and human heart through public databases. Moreover, we used bioinformatics analysis combined with network structuring to deduce the underlying mechanism of abnormal cardiac function and myocardial injury in patients with COVID-19. The findings indicated that ACE2 is not only a SARS-CoV-2 receptor but also a key component in the pathogenesis of COVID-19-related cardiac injury. We highlighted the need for clinicians to pay attention to SARS-CoV-2-associated cardiac complications and provide new evidence for clinical therapeutics.
Methods
mRNA and protein expression data in human tissue
The distribution of ACE2 in healthy human organs, along with the clinical characteristics were derived from the Genotype-Tissue Expression (GTEx) project, which includes healthy human tissue mRNA expression data from post-mortem donors.
All of the GTEx data were downloaded from the Human Protein Atlas (14) (HPA, https://www.proteinatlas.org/), and the log2 scale was used for expression calculation and visualization. The protein expression level of normal tissues was identified using immunohistochemistry. Immunohistochemistry images and antibody annotations were also obtained from HPA. The ACE2 expression data in the normal and cardiomyopathy heart were collected in the Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database (15) (GSE3585 and GSE26887).
mRNA expression data in mouse heart
The expression data in mice were downloaded from the Expression Atlas of the European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI, https://www.ebi.ac.uk/) database (16). The expression atlas provides gene expression data across different biological conditions and developmental stages. Data sets E-MTAB-6798 and E-GEOD-49906 were selected to explore changes of ACE2 expression during postnatal mouse heart development. The heatmap was plotted using GraphPad Prism 8.
Gene correlation analysis
Gene correlation analysis was performed using the web-based tool, Gene Expression Profiling Interactive Analysis (17) (GEPIA, http://gepia.cancer-pku.cn/). GEPIA provides pair gene correlation based on the Cancer Genome Atlas (TCGA) and GTEx database. In this study, only the left ventricle gene expression set of GTEx was selected for analysis. Spearman rank correlation was used to determine the co-expression between ACE2 and virus-related genes.
Gene ontology (GO) analysis
Search Tool for the Retrieval of Interacting Genes/Proteins (STRING, https://string-db.org/) is a website predicting the interaction partners of input protein according to a combined score (18). The 30 proteins that had the highest combined score with ACE2 in STRING were selected for further analysis. The biological processes of ACE2 and its 30 predicted partners were then explored through a GO enrichment analysis (19) using Metascape (https://metascape.org/) (20), and the top 20 enriched biological process terms were displayed.
Protein-protein interaction (PPI) network with virus-related proteins
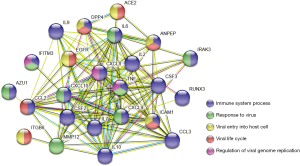
To explore the possible mechanisms of ACE2 in COVID-19, 23 proteins involved in virus biological activities or clinical progression were selected based on previous reports. Then, a PPI network of ACE2 and 23 virus-related proteins was constructed using String, where the color of the node indicated which GO terms the protein was enriched by, and the color of the edge stood for the type of interaction evidence.
Statistical analysis
The statistical analysis was carried out using GraphPad Prism 8 (GraphPad Software Inc., San Diego, CA, USA). The normality of data was determined by Shapiro-Wilk test. If data were normally distributed, the Student’s t-test was used to determine whether the mean expression of two groups was the same, and one-way analysis of variance (ANOVA) for three or more groups. If the data were not normally distributed, the non-parametric Mann-Whitney test was used to compare the difference between two groups, and Kruskal-Wallis test for three or more groups. To adjust the P value in multiple testing, the two-stage step-up method of Benjamini, Krieger, and Yekutieli was applied as post-test to control the false discovery rate. The gene correlations were estimated by Spearman’s rank correlation coefficients. All tests were two-sided and were significant when both P and q<0.05.
Results
ACE2 expression levels in different types of human organs
To determine the expression of ACE2 in normal human heart tissue, the ACE2 mRNA levels in different organs or tissues were analyzed using the GTEx database. We first overviewed the ACE2 mRNA expression pattern in healthy human major organs (Figure 1). The results demonstrated that the mRNA level of ACE2 in cardiac tissues including the left ventricle and atrial appendage was significantly higher than that in the lung (P<0.0001, q<0.0001; Figure 1). ACE2 mRNA expression has no difference between the left ventricle and the atrial appendage (P=0.5469, q=0.1618; Figure 1). Thus, we used left ventricle expression data to explore the role of ACE2 in the subsequent study.
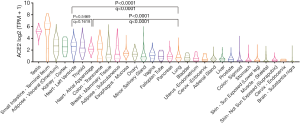
mRNA and protein expression of ACE2 in the human heart
To estimate the cardiac injury risk mediated by the different coronavirus infections, we investigated the expression of the corresponding coronavirus receptors in cardiac tissues. As shown in Figure 2A, ACE2 mRNA was significantly higher than aminopeptidase N (ANPEP, the receptor for human coronavirus 229E, HCoV-229E) and dipeptidyl peptidase 4 (DPP4, the receptor for Middle East respiratory syndrome coronavirus, MERS-CoV) in healthy left ventricle tissues (P<0.0001, q<0.0001). Next, we examined the relationship between ACE2 expression and the clinical characteristics, but observed no significant difference in ACE2 expression between age groups and gender groups (Figure 2A). To determine the protein expression of ACE2 in the human heart, we downloaded immunohistochemistry images from the Human Protein Atlas. As shown in Figure 2B, the expression level of ACE2 protein was markedly elevated in the heart and mainly localized in the human cardiomyocytes, but no positive staining of ANPEP or DPP4 protein was detected. Furthermore, we compared the mRNA expression of ACE2 between healthy and cardiomyopathic heart samples. The clinical characteristics of GEO cohorts are listed inTable S1. The results of both data sets showed that the mRNA expression of ACE2 was markedly increased in cardiomyopathic heart samples compared with that in healthy heart samples (P=0.0048, P=0.0076; Figure 2C), which suggests a higher risk of direct cardiac damage in cardiomyopathy patients after SARS-CoV-2 infection.
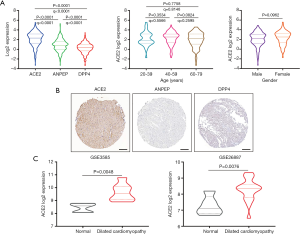
Expression level of ACE2 is upregulated during mouse heart development
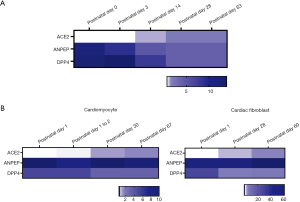
To explore ACE2 expression at different developmental stages of the heart, we compared the expression level of ACE2 in mouse heart, together with coronavirus receptors ANPEP and DPP4 at five points in time after birth in data set E-MTAB-6798. The heatmap showed that the expression of ACE2 in the heart markedly increased during mouse growth, whereas the expression of ANPEP and DPP4 presented the opposite tendency (Figure 3A). To further evaluate the change in ACE2 expression in different cells of cardiac tissues, the mRNA expression of ACE2 in mouse cardiomyocytes and cardiac fibroblasts was analyzed using data set E-GEOD-49906. We found that ACE2 expression continuously increased with development in both cardiomyocyte and cardiac fibroblasts (Figure 3B), which was consistent with the result of the whole mouse heart. However, the expression of ANPEP and DPP4 revealed no trend in the same analysis (Figure 3B). These findings showed that the expression of ACE2 in the heart was enhanced with an increase of age.
Correlation analysis between ACE2 expression and virus-related genes
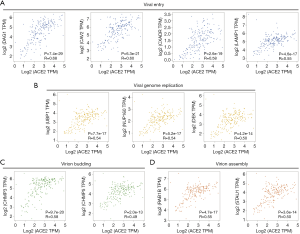
To investigate the relationship between ACE2 expression and viral infection, we selected gene markers that participate in the whole viral process. These gene markers were put into GEPIA and performed pair gene correlation with ACE2. The Spearman correlations analysis demonstrated that ACE2 expression was positively correlated with many virus-related genes, including entering into host cells, viral genome replication, virion assembly, and budding (Figure 4). Notably, we found that the virus-related genes involved in entering into the host cell, such as dystroglycan 1 (DAG1), caveolin 2 (CAV2), coxsackie virus and adenovirus receptor (CXADR), and lysosomal-associated membrane protein 1 (LAMP1), had strong correlations with ACE2 (Figure 4A). Furthermore, ACE mRNA levels were more strongly correlated with virion assembly and budding-related genes containing charged multivesicular body protein 3 (CHMP3) and Ras-related protein Rab-1B (RAB1B) (Figure 4C,D).
GO analysis
We used the STRING website to explore the interaction relationship of ACE2, and the top 30 predicted proteins as sorted by scores were acquired for further analysis (Table 1). The list included several proteins that regulate viral activities, including DPP4, adrenergic beta receptor kinase 1 (ADRBK1), and epidermal growth factor receptor (EGFR). To annotate the roles of 31 proteins, GO enrichment analysis was performed using Metascape. The top 20 significant GO enrichment terms of biological processes are illustrated in Figure 5. The results of GO analysis showed that ACE2-associated proteins were mainly enriched in the regulation of cardiovascular functions, which was consistent with its original roles (Figure 5). Interestingly, biological processes, such as cytokine production, viral entry into the host cell, and neutrophil-mediated immunity, were significantly enriched among ACE2-associated proteins (Figure 5).
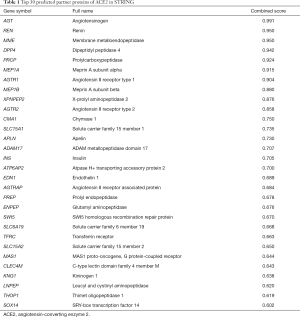
Full table

PPI network with virus-related proteins
To better understand the underlying mechanisms of ACE2 in virus-induced heart damage, we reviewed the latest literature and selected 23 proteins consisting of two parts: viral process-related proteins, and proinflammatory cytokines or chemokines that markedly increased in COVID-19 patients’ plasma (6). All of the 24 proteins, including ACE2, were put into STRING for network construction. As shown in Figure 6, ACE2 had direct interaction with interleukin 6 (IL6), which is a pleiotropic cytokine involved in regulating acute inflammatory responses. Moreover, the network displayed that ACE2 could cross-talk with the other two coronavirus receptors ANPEP or DPP4, leading to the production of more proinflammatory factors and the regulation of other virus-related proteins as well (Figure 6).
Discussion
Cardiac injury is a fatal complication following COVID-19. According to a clinical report containing 138 hospitalized patients, 7.2% of patients developed acute cardiac injury, and the patients admitted to the ICU were more likely to suffer cardiac injury compared with the mild cases (5). ACE2 proved to be the major receptor for SARS-CoV-2; its distribution in heart tissue could encourage us to take the corresponding measures to prevent cardiac injury in patients with COVID-19. In this study, we first used the public database to analyze the ACE2 expression in the heart of normal people, and found a higher mRNA level of ACE2 than that in lung tissue, which was mostly expressed in the left ventricle. The results suggested that the heart is also potentially vulnerable to the SARS-CoV-2 infection.
It is worth noting that severe and fatal cases were strongly associated with older age and comorbidities. Only 2.4% of cases were under 19 years of age, and most of them had a favorable prognosis, while only a few eventually developed into critical cases (21). The total CFR of confirmed cases was 3.8%, but the CFR in patients with cardiovascular disease reached up to 13.2% (21), which was higher than that in other comorbidities including diabetes, hypertension, chronic respiratory diseases, and cancers. Based on the above clinical features in COVID-19, we explored disparities in ACE2 expression in two populations: children and cardiovascular patients. In this study, the time course of the postnatal mouse model showed that ACE2 expression in the heart tissue increased with cardiac development, which suggested that SARS-CoV-2 may have less cardiac impact on children than on adults. We then compared the expression of ACE2 in underlying cardiovascular disease and healthy heart tissue. The findings demonstrated that patients with underlying cardiovascular disease had higher ACE2 expression in the heart tissue. This result was consistent with the study that ACE2 played a protective role in the cardiovascular system after myocardial infarction by counter-regulatory mechanisms (22). Hence, cardiovascular disease patients are likely at increased risk of cardiac injury after SARS-CoV-2 infection.
The mechanisms of cardiac injury in patients with COVID-19 remain unclear. In the first autopsy of a patient with COVID-19, minor inflammatory mononuclear infiltration was found in the heart tissue, but the result provided no evidence of direct damage (23). Moreover, given the absence of more autopsy reports, we reviewed published literature about the cardiac implications of SARS. In autopsy research containing 20 SARS patients (24), SARS-CoV RNA was found in the heart tissue of 35% of infected patients, along with significant macrophage infiltration and myocardial injury. Considering that SARS-CoV and SARS-CoV-2 share ACE2 as the major receptor (10), we hypothesized that the same damage may occur in patients with COVID-19. In this study, we compared the expression of ACE2 with the other coronavirus receptors, such as ANPEP and DPP4 in the human heart tissue. The results showed that ACE2 expression was significantly higher than other coronavirus receptors, indicating that patients with COVID-19 may face a higher risk of heart injury. Besides, ACE2 was strongly co-expressed with genes necessary for viral activities, especially the process of entering into the host cell. Therefore, it seems that SARS-CoV-2 has evolved to hijack cardiomyocytes for its reproduction and transmission mediated by ACE2.
During the outbreak of COVID-19, cytokine storm is considered to accelerate the progression of severe cases and cause many deaths (6). The cytokine storm is an exaggerated and often lethal immune response, proinflammatory cytokines are continuously produced and finally come to a vicious cycle. In previous reports, the high expression of ACE2 was also observed in infiltrating mononuclear cells after cardiac injury (22), indicating that ACE2 plays a potential role in the inflammatory response. Therefore, we constructed the PPI network, including virus-related proteins and proinflammatory factors, to explore the relationship between ACE2 and the cytokine storm. We collected these proteins from three sources: (I) the proteins were annotated to affect the virus entering and lung injury after infection from UniProt website (www.uniprot.org). (II) The proteins were correlated to ACE2 expression and participated in the anti-viral process (25). (III) Inflammatory cytokines were elevated in ICU and non-ICU patients with COVID-19, which might lead to more severe clinical progression (6). The network revealed that ACE2 directly interacted with IL6 and indirectly affected other proinflammatory factors. IL6 and granulocyte-macrophage colony stimulating factor (GM-CSF) are believed to be the key inflammatory factors that trigger the cytokine storm in patients with COVID-19 (26). In the latest “Diagnostic and treatment protocol for COVID-19”, the progressive increase of IL6 was used as a clinical warning indicator for the deterioration of patients with COVID-19. Moreover, for the first time, Tocilizumab was recommended in the treatment of severe and critical cases (27). Previous literature has shown that ACE2 may act as an upstream factor of IL6 and regulate its expression (25). Thus, besides acting as the receptor for SARS-CoV-2, ACE2 may participate in the regulation of cardiac inflammation in the cytokine storm.
This study had several limitations that may cause analysis bias. The low-risk conclusion on children was drawn from the postnatal mouse model and therefore may not be reliably extrapolated to humans. It has also been reported that the expression of ACE2 is associated with racial differences, but we did not include race as a factor in this study, as such information was unavailable in available data sets. This study was mostly based on data from a healthy human public database, but considering the unique pathological conditions in patients with COVID-19, the results still need to be confirmed through further clinical observations.
Conclusions
In this study, we applied multiple public databases to investigate the expression pattern of ACE2 in cardiac tissue and further explored its potential roles in cardiac injury during SARS-CoV-2 outbreak. Our findings suggested that SARS-CoV-2 may attack the heart tissue directly by binding to the ACE2 receptor. Meanwhile, ACE2 could contribute to multiple viral processes in myocardial cells and participate in the regulation of the cardiac inflammatory response following SARS-CoV-2 infection. Given the results of ACE2 expression pattern among populations, it is reasonable to apply additional precautions and strengthen the monitoring of heart function and myocardial injury biomarkers in patients with COVID-19 who experience cardiovascular disease complications. Although our study showed that cardiac risk was relatively low in children and uncomplicated cases, potential cardiac injury should be of particular concern in follow-ups. Keeping vigilance is extremely important before more information is available.
Acknowledgments
The authors would like to thank the public databases for the availability of the data.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-2755). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Ethical approval was waived because we used only publicly available data in this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Yu CM, Wong RSM, Wu EB, et al. Cardiovascular complications of severe acute respiratory syndrome. Postgrad Med J 2006;82:140-4. [Crossref] [PubMed]
- Alhogbani T. Acute myocarditis associated with novel Middle east respiratory syndrome coronavirus. Ann Saudi Med 2016;36:78-80. [Crossref] [PubMed]
- Oudit GY, Kassiri Z, Jiang C, et al. SARS-coronavirus modulation of myocardial ACE2 expression and inflammation in patients with SARS. Eur J Clin Invest 2009;39:618-25. [Crossref] [PubMed]
- Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 2020;395:565-74. [Crossref] [PubMed]
- Wang D, Hu B, Hu C, et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020;323:1061-9. [Crossref] [PubMed]
- Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506. [Crossref] [PubMed]
- Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 2020;395:1054-62. [Crossref] [PubMed]
- Wu C, Hu X, Song J. Heart injury signs are associated with higher and earlier mortality in coronavirus disease 2019 (COVID-19). medRxiv 2020.
- Hui H, Zhang Y, Yang X, et al. Clinical and radiographic features of cardiac injury in patients with 2019 novel coronavirus pneumonia. medRxiv 2020.
- Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020;181:271-280.e8. [Crossref] [PubMed]
- Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020;367:1260-3. [Crossref] [PubMed]
- Zhang H, Kang Z, Gong H, et al. The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes. bioRxiv 2020.
- Fan C, Li K, Ding Y, et al. ACE2 Expression in Kidney and Testis May Cause Kidney and Testis Damage After 2019-nCoV Infection. medRxiv 2020.
- Uhlén M, Fagerberg L, Hallstrm BM, et al. Tissue-based map of the human proteome. Science 2015;347:1260419. [Crossref] [PubMed]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30:207-10. [Crossref] [PubMed]
- Petryszak R, Fonseca NA, Fllgrabe A, et al. The RNASeq-er API-a gateway to systematically updated analysis of public RNA-seq data. Bioinformatics 2017;33:2218-20. [Crossref] [PubMed]
- Tang Z, Li C, Kang B, et al. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98-W102. [Crossref] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607-13. [Crossref] [PubMed]
- Ashburner M, Ball CA, Blake JA, et al. Gene Ontology: tool for the unification of biology. Nat Genet 2000;25:25-9. [Crossref] [PubMed]
- Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10:1523. [Crossref] [PubMed]
- Report of the WHO-CHINA joint mission on COVID-19. National Health Commission of the People’s Republic of China 2020.
- Burrell LM, Risvanis J, Kubota E, et al. Myocardial infarction increases ACE2 expression in rat and humans. Eur Heart J 2005;26:369-75. [Crossref] [PubMed]
- Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med 2020;8:420-2. [Crossref] [PubMed]
- Guo Y, Korteweg C, McNutt MA, et al. Pathogenetic mechanisms of severe acute respiratory syndrome. Virus Res 2008;133:4-12. [Crossref] [PubMed]
- Chen L, Zhong L. Lung adenocarcinoma patients own higher risk of SARS- CoV-2 infection. Preprints 2020.
- Zhou Y, Fu B, Zheng X, et al. Aberrant pathogenic GM-CSF+ T cells and inflammatory CD14+CD16+ monocytes 1in severe pulmonary syndrome patients of a new coronavirus. bioRxiv 2020.
- Diagnostic and treatment protocol for COVID-19 (trial version 7). National Health Commission of the People’s Republic of China2020.


