Comprehensive bioinformatics analysis of the TP53 signaling pathway in Wilms’ tumor
Introduction
Wilms’ tumor (WT), which is also known as nephroblastoma, is an embryonal tumor derived from the metanephros. WT, which accounts for 90% of pediatric renal tumors and 7% of all pediatric cancers (1), mainly affects children under the age of 10 years.
WT is caused by the aberrant development of the kidney, and a number of genetic aberrations have been observed during its development. The aberrant expression of different genes may lead to different outcomes. Among the genes with aberrant expression in WT, Wilms’ tumor 1 (WT1) is associated with stromal histology (2). MYCN is also an adverse biomarker and is significantly related to anaplastic histology (3). Meanwhile, MLLT1 gene aberration may be associated with younger age at diagnosis (4). Tumor protein 53 (TP53), which has been reported to be related to poor outcomes, is rarely observed in tumors without diffuse anaplasia (5). For most patients with WT, the treatment has been well classified. However, the effective treatment of children with bilateral, relapsed, high-risk tumors, as well those with diffuse anaplastic WT (DAWT), is still challenging, and these patients often have poor outcomes (6).
WT is clinically and biologically heterogenic. Fundamental knowledge of Wilms tumor-related genes via analyzing the Wilms tumor predisposition syndromes patients, identifying the mutated Wilms' tumor suppressor 1 (WT1) gene as the driver factor for tumorigenesis (7). Further analysis of WT1 showed that WT1 is one of most important regulators of podocyte-specific transcriptome (8). With the further understanding the tumor-related genes in WT. There were several gene changes such as CTNNB1, AMER1, IGF2, and MLLT1 that played a key role in WT (4,9-11). These genetics changes in WT that further approve the WT is a typical genetic disease. Among these gene aberrations, TP53 is one of the most commonly mutated gene in WT. The TP53 protein plays a fundamental and multifaceted role as a tumor suppressor in tumorigenesis, tumor progression, and therapeutic response. There were several modification types and regulation of TP53 such as phosphorylation, acetylation, ubiquitination. However, the function of these modification of TP53 still obscure. To date, the changes of TP53 signaling pathway have been observed in many cancer types. Even for several cancer types that mutated TP53 or TP53 signaling pathway had severed as the therapy targets for multiple cancer types including multiple myeloma, osteosarcoma, bladder cancer, and myelodysplastic syndromes (12-15). The drugs and small molecules that targets TP53 or TP53 signaling pathway including the most widely understood were PRIMA-1 [2,2-bis(hydroxymethyl)quinuclidin-3-one or APR-017] and APR-246 [2-(hydroxymethyl)-2-(methoxymethyl)] that induced the cancer cell apoptosis (16). The TP53 may be the important regulator of apoptosis, senescence, and metabolism for cancer (6,17,18). From these, the TP53 may sever as a key role for WT as well.
The precise biological mechanism underlying the function of TP53 as a tumor suppressor has yet to be well defined in WT. The tumor-suppressive function of the genes involved in the TP53 signaling pathway is of profound importance to improving our understanding of cancer biology, including tumorigenesis, tumor progression, and drug resistance, as well as to developing more effective cancer therapies. The present study aimed to comprehensively analyze the prognostic role and biological functions of the TP53 signaling pathway in WT, and to explore the regulators of the pathway.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-6047).
Methods
Statistical analysis
The gene expression, survival analysis, and the protein-protein network relations P value <0.05 is considered a significant difference in our study.
Analysis of the TP53 signaling pathway in WT using cBioPortal
The cBioPortal for Cancer Genomics (http://cbioportal.org) allows users to explore, visualize, and analyze multidimensional cancer genomics data online (19). We used cBioPortal to analyze the mutation types, frequencies, and locations of genes involved in the TP53 signaling pathway, as well as the relationships between these genes, and the clinical features and prognosis of WT.
Analysis of TP53 signaling pathway gene expression levels using Oncomine
The Oncomine database (https://www.oncomine.org/resource/login.html) contains information on the genes, pathways, and networks deregulated across 18,000 cancer gene expression microarrays, spanning the majority of cancer types and subtypes (20). We used Oncomine to analyze the expression of TP53 signaling pathway genes in WT.
Analysis of microRNAs (miRNAs) targeting the TP53 signaling pathway
miRNAs are important regulators of gene expression at the transcriptional and post-transcriptional levels. We used the miRTarBase (21) and miWalk databases (22) to investigate the miRNAs that target genes involved in the TP53 signaling pathway. A Venn diagram was produced to identify the miRNAs that co-target TP53 signaling pathway genes.
Protein-protein network analysis via GeneMANIA
GeneMANIA is an online tool that enables users to construct protein-protein interaction (PPI) networks and to predict the potential functions of defined genes (23). GeneMANIA allows the relationships between genes, including co-expression and physical interaction, to be visualized. We used GeneMANIA to construct the PPI network of TP53 signaling pathway-related genes.
ChIP-X enrichment analysis 3 (ChEA3)
ChEA3 (https://amp.pharm.mssm.edu/ChEA3) is an online enrichment analysis tool that ranks the TFs associated with genes (24). We used ChEA3 to find TF targets, and to annotate the main biological functions of the genes involved in the TP53 signaling pathway.
Human Protein Atlas (HPA) analysis
The HPA (http://www.proteinatlas.org) aims to map all of the human proteins in cells, tissues, and organs by integrating various omic technologies, including antibody-based imaging, mass spectrometry-based proteomics, transcriptomics, and systems biology (25). We used the HPA database to investigate the subcellular localization and siRNA inhibition of TP53 expression.
Analysis of the potential drug targets of TP53 using Cancer Therapeutics Response Portal (CTRP)
To investigate the drugs that could potentially be used to target the TP53 signaling pathway, we used the CTRP database, which links the genetic, lineage, and other cellular features of cancer cell lines to small-molecule sensitivity with the aim of supporting the discovery of potential drugs or small molecules for cancer therapy (26).
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Expression analysis of genes in the TP53 signaling pathway
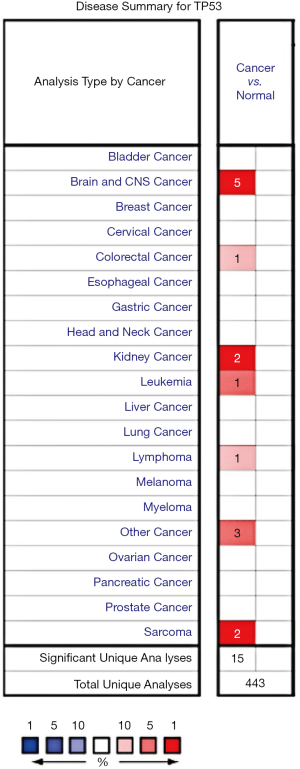
The Oncomine database was used to investigate the TP53 mRNA expression levels in various cancers. The results showed that TP53 is overexpressed in cancers of the brain and the central nervous system, as well as in colorectal cancer, kidney cancer, leukemia, lymphoma, and sarcoma (Figure 1).
Next, we analyzed the TP53 expression levels specifically in WT. A significant difference existed between the levels of TP53 expression in WT samples and normal samples in the Cutcliffe Renal dataset [P value =0.010, fold change (FC) =1.976]; however, TP53 expression was not significantly different between tumor samples and normal samples in the Yusenko Renal dataset (P value =0.094, FC =1.984) (Figure 2A,B).
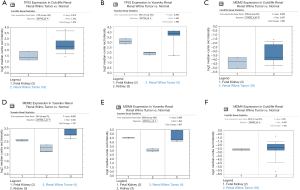
Subsequently, we also explored the expression levels of the genes involved in the TP53 signaling pathway. The results showed that MDM2 was significantly overexpressed in the Yusenko Renal dataset (P value =0.05, FC =1.747), but not in the Cutcliffe Renal dataset (P value =0.058, FC =1.955) (Figure 2C,D). The expression levels of MDM4 showed no significant difference in the Yusenko Renal dataset (P value =0.055, FC =1.837) or in the the Cutcliffe Renal dataset (P value =0.501, FC =−1.001) (Figure 2E,F). These results indicated that the TP53 signaling pathway genes are differentially expressed in WT and, therefore, may serve an important role in the tumorigenesis and prognosis of WT.
Analysis of gene mutation types, locations, structures, co-expression, and effectors of expression
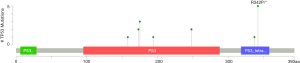
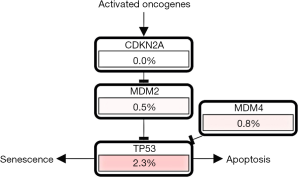
Different mutation types may have distinct biological functions in various cancers, and the co-expression of genes may also affect gene functions. Using the cBioPortal database, we explored the location and types of mutation, as well as co-expression, among genes related to the TP53 signaling pathway. Among these genes, TP53 (2.3%) was the most frequently aberrant gene in WT, followed by MDM4 (0.8%), and then MDM2 and TP53BP1 (0.2%). The most common types of mutation in TP53 were missense and deep deception, while amplification was the most common mutation in MDM2, MDM4, and TP53BP1 (Figure 3A,B). The three-dimensional structure of TP53 is shown in Figure 3C.

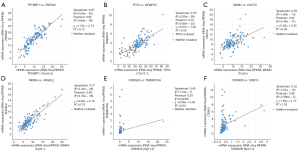
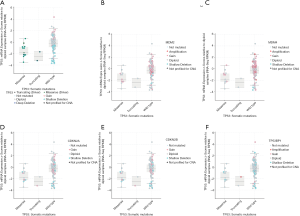
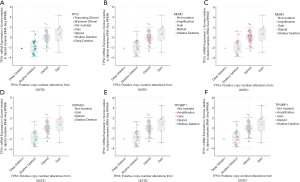
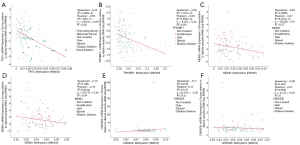
The location of mutations in TP53 are shown in Figure 4. The significant co-expressed genes in the TP53 signaling pathway included ZNF609 (zinc finger protein 609), WRAP53 (WD40-encoding RNA antisense to p53), CNOT2 (CC chemokine receptor 4-negative regulator of transcription 2), ANGEL2, TMEM255A, and CDH13 (cadherin 13) (Figure 5). Further, we investigated the relationships of the mutation types (Figure 6), copy number alterations (CAN, Figure 7), and methylation (Figure 8) with the expression levels of defined genes. The gene expression levels were negatively correlated with methylation, but the CNA and mutated type did not have a significant influence. Figure 9 shows the relationship between the genes in the TP53 signaling pathway.
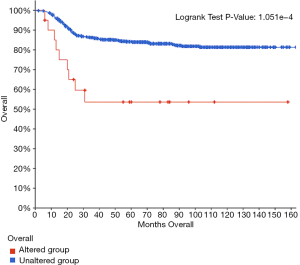
Survival analysis
To investigate the prognostic role of the aberrant TP53 signaling pathway in WT, we used cBioPortal to perform survival curve analysis. The results indicated that the group with alterations had shorter overall survival than the group without alterations (Figure 10), suggesting that the TP53 signaling pathway plays an important role in WT.
miRNA target genes in the TP53 signaling pathway
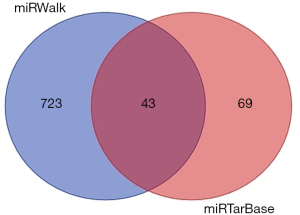
MiRTarBase, and miWalk were used to identify miRNA target genes in the TP53 signaling pathway. Further, we used the miWalk dataset to construct a miRNA network (Figure 11). Finally, a Venn diagram was produced to explore the co-targeting miRNAs of defined genes (Figure 12). There were 69 and 723 miRNA targets of TP53 from the miRTarBase and miWalk datasets, respectively. There were 43 co-targeting miRNAs including hsa-miR-17-5p, hsa-miR-5196-3p, hsa-miR-18a-5p, hsa-miR-6780b-5p, hsa-miR-125a-5p, hsa-miR-6797-5p, hsa-miR-6825-5p, hsa-miR-8071, hsa-miR-6751-5p, hsa-miR-19b-3p, hsa-miR-30a-5p, hsa-miR-6880-5p, hsa-miR-6129, hsa-miR-6803-5p, hsa-miR-7150, hsa-miR-1249-5p, hsa-miR-92a-3p, hsa-miR-92a-1-5p, hsa-miR-622, hsa-miR-194-3p, hsa-miR-149-3p, hsa-miR-222-3p, hsa-miR-6835-5p, hsa-miR-4650-5p, hsa-miR-324-5p, hsa-miR-6752-5p, hsa-miR-4271, hsa-miR-612, hsa-miR-5193, hsa-miR-4651, hsa-miR-7110-5p, hsa-miR-6722-3p, hsa-miR-6127, hsa-miR-150-5p, hsa-miR-6133, hsa-miR-4510, hsa-miR-30e-5p, hsa-miR-6749-3p, hsa-miR-6785-5p, hsa-miR-485-5p, hsa-miR-5003-5p, hsa-miR-3529-3p, and hsa-miR-4736.
TF target analysis
TFs are important regulators of gene expression and transduction. Therefore, we used ChEA3 to analyze the TF targets of TP53 signaling pathway genes, to further explore their distribution and biological functions. The results revealed that the functions of the TF targets included immune response, organ development, DNA transcription, and lipid biosynthesis (Figure 13A). The TFs were found to be distributed in various tissues, such as the liver, brain, vessels, and adipose tissue (Figure 13B). The top 10 TFs included SERBF1, ZNF274, TEAD4, EGR1, and CTCF (Figure 13C).

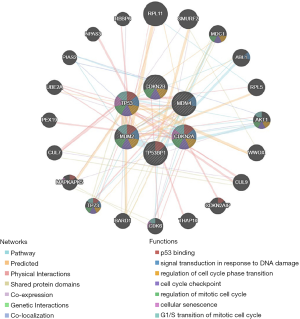
GeneMANIA analysis
The relationships between the TP53 signaling pathway genes and the other genes were explored by constructing a PPI network using GeneMANIA. The functions in the PPI network included p53 binding, signal transduction in response to DNA damage, regulation of cell cycle phase transition, cell cycle checkpoint, regulation of mitotic cell cycle, cellular senescence, and G1/S transition of the mitotic cell cycle. Moreover, the TP53 signaling pathway genes were significantly related to TP73, AKT1, and MDC1 (Figure 14).
HPA analysis
TP53 subcellular localization was explored using the HPA dataset for immunofluorescence staining of TP53. The results showed that the TP53 is mainly expressed in the nucleus (Figure 15A) and that siRNA could decrease the levels of TP53 expression (Figure 15B). These results indicated that regulating TP53 expression could be a potential approach to treating cancer.
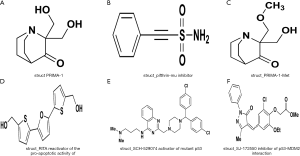
CTRP analysis
Finally, we used CTRP to explore the potential drugs and small molecules that target TP53 signaling pathway genes. The results showed that six small molecules, including PRIMA-1, pifithrin-mu, PRIMA-1-Met, RITA, SJ-172550, and SCH-529074, target the TP53 signaling pathway (Figure 16). Among the small molecules, RITA and SCH-529074 were activators of TP53, while pifithrin-mu and SCH-529074 were inhibitors.
Discussion
Despite the development of therapy protocols, relapse and several subtypes still present considerable challenges in the clinical treatment of WT. Despite more than 30 years of various studies and an expansive body of literature, the precise molecular mechanisms underlying tumorigenesis, tumor progression, drug resistance, and heterogeneity in WT have yet to be fully elucidated. Genetic changes play an important role in tumorigenesis, tumor progression, and therapeutic response in multiple cancers. Therefore, gene aberrations may serve as biomarkers, or even as therapeutic targets, for some malignancies. In WT, aberrant changes in the TP53 signaling pathway are the most common. Genetic changes in the TP53 signaling pathway have been shown to lead to poor prognosis in cancer patients (27,28). However, the underlying biological functions of the TP53 signaling pathway remain to be fully understood. The potential mechanisms of TP53 signaling pathway changes that lead to an adverse outcome of WT patients may include the changes lead to inactivate or inhibit normal TP53 signaling pathway transduction. So, the functions of several targets TP53 signaling pathway genes that restore the function of TP53. TP53 signaling pathway to therapy cancer. Due to the mutation frequencies and the important role of the TP53 signaling pathway in various cancers, we investigated the expression levels of its related genes in WT samples, with the aim of analyzing their prognostic role in the disease. The results showed TP53 and MDM2 were highly expressed in tumor samples compared with normal samples, suggesting that aberrant changes in TP53 signaling pathway genes can lead to the poor outcomes of WT patients. Tumorigenesis, tumor progression, and therapy resistance are not determined by a single gene mutation; instead, they are caused by several important genetic changes. The co-expressed genes involved in the TP53 signaling pathway included ZNF609, WRAP53, CNOT2, ANGEL2, TMEM255A, and CDH13; however, these genes do not have a significant role in the prognosis of WT. Wu et al.’s study revealed ZNF609 to be a promoter of migration in colon cancer cells (29), and it has also been suggested as a driver factor in gastric cancer (30). Silwal-Pandit et al.’s study showed that WRAP53 could serve as a biomarker for diagnosing and predicting the prognosis of breast cancer patients, it could reverse the prognosis of breast cancer patients with TP53 mutations (31). Sohn et al. showed that CNOT2 could accelerate breast cancer cell proliferation and angiogenesis via VEGFR (vascular endothelial growth factor receptor) signaling (32). Higher expression of CDH13 can reverse the resistance of non-small cell lung cancer cells (NSCLCs) to cisplatin (33). These studies indicate that the TP53 signaling pathway and its co-expressed genes may play a key role in WT. Unfortunately, the role of co-expressed genes in WT has not been fully explored to date. Further research should focus on the relationship between the TP53 signaling pathway and gene co-expression, and explore the role of co-expressed genes in WT.
Gene expression and cell signaling transduction may be impacted by several factors. The most important factors include miRNAs and TFs. Our miRNA target analysis showed that there are a number of miRNA targets in the TP53 signaling pathway. Hsa-miR-485-5p can reverse epithelial-mesenchymal transition (EMT) to promote cisplatin-induced death of oral squamous cell carcinoma (OSCC) cells (34). Hsa-miR-150-5p has been observed to be upregulated in chronic lymphocytic leukemia, although its function is still unknown (35). Hsa-miR-5193 was found to suppress motif-containing protein 11 (TRIM11) to inhibit the progression of prostate cancer (36), while Hsa-miR-612 was shown to inhibit the growth of colon cancer and hepatocellular carcinoma (37,38). Has-miR-212-3p has been detected in human glioma (39). Furthermore, other miRNAs have been observed in multiple cancers (40-43). The results of these studies indicate that the miRNA targets of TP53 play a significant role in tumor progression and resistance to therapy; however, the functions of these targets in WT had not been classified, and the relationship between these miRNAs and TP53 signaling pathway was still unclear. The TF targets of TP53 include KDM5B, NR2F2, CTCF, ZNF274, TEAD4, EGR1, IRF3, and SREBF1. NR2F2 promotes tumor growth factor (TGF)-β-dependent epithelial–mesenchymal transition of colon cancer cells by inhibiting Smad7 expression (44). This process is likely to be involved in the progression of multiple solid cancers. Furthermore, NR2F2 overexpression has been shown to indicate poor prognosis in gastric cancer (45). KDM5B has been detected in multiple cancers, including breast cancer, melanoma, NSCLC, and gastric cancer, and KDM5B overexpression can serve as an adverse biomarker for these cancers (46-49). TEAD4 also has an oncogenic role in gastric cancer, and its inhibition was shown to produce an anticancer effect (50). IRF3 is a positive regulator of YAP (parts of hippo signaling pathway) and a the driver in several malignancies (51). Sterol regulatory element-binding transcription factor 1 (SREBF1), which is regulated by the AR/mTOR nuclear axis (52), has been shown to be a possible regulator of the metabolic process in prostate cancer. These results indicate that the TF targets of TP53 and its signaling pathway genes serve as the drivers of tumorigenesis and tumor progression. However, EGR1 has been demonstrated to be a protector in NSCLC, with its inhibition promoting the invasion and proliferation of cancer cells (53). Regrettably, the TF targets of TP53 are still unclear in WT.
The ultimate goal of many studies is to find more effective treatment protocols to lengthen the lifespan of, and to even cure, patients. Therefore, we used the HPA dataset to explore the location of TP53 expression. The results showed that TP53 expression is localized in the nucleus and could be inhibited by siRNA. Recent studies have explored whether siRNAs can be used to treat cancer. Singh et al. showed that siRNA could be used to deliver anticancer drugs and to inhibit genes at the post-transcriptional level (54). The effectiveness of anticancer drugs can be considerably influenced by their absorption, distribution, metabolism, and excretion. TP53 expressed in the nucleus can lead to the development of resistance to some anticancer drugs (55). TP53 is the most mutated gene in multiple cancers and the reactivation of its inactive mutated form represents a possible target for antitumor therapy. Further, we used the CRTP dataset to explore the potential target drugs and small molecules of the TP53 signaling pathway. The results showed that PRIMA-1, pifithrin-mu, PRIMA-1-Met, RITA, SJ-172550, and SCH-529074. PRIMA-1, and Prima-1-Met were activators of TP53. Activation of TP53 can induce cancer cell apoptosis (56). Pifithrin-mu was shown to inhibit heat shock protein 70 (HSP70) and could be combined with other chemotherapy drugs to improve their efficacy (57). Pifithrin-mu also served as the anticancer molecule of NSCLC (58). RITA is a small molecule that blocks TP53-MDM2 interaction, thereby reactivating TP53 in tumors to induce tumor cell apoptosis. RITA combined with 3-methyladenine (3-MA) can be used to treat chemoresistance in head and neck cancer by inhibiting the autophagy and antioxidant systems (59). We used the cBioPortal dataset to explore the TP53 signaling pathway. The results showed that TP53 also interacted with MDM4. SJ-172550 is an inhibitor of the interaction of TP53 and MDM4 (60), while SCH-529074 can reactivate mutated TP53 to suppress NSCLC cell growth (61).
These findings indicate that mutated TP53 loses its tumor-suppressive effect, while reactivation of TP53 can restore its functions, thus suggesting its anticancer effect. The aberrant TP53 signaling pathway is potentially the driver of several cancers, and blocking the interaction TP53 and the signaling pathway parts can inhibit the growth and progression of cancer.
In the present study, we comprehensively analyzed the TP53 signaling pathway and preliminarily explored the biological functions. Our results showed that several TP53 signaling pathway genes were differentially expressed in WT samples compared to normal tissue samples. TP53 is the most frequently mutated gene in the TP53 pathway in WT, and TP53 mutations were an indicator of poor prognosis. The functions of TP53 signaling pathway genes included p53 binding, signal transduction in response to DNA damage, regulation of cell cycle phase transition, cell cycle checkpoint, regulation of mitotic cell cycle, cellular senescence, and G1/S transition of the mitotic cell cycle. The important miRNAs (miR-485-3p) targets and TFs (NR2F2 and KDM5B) of TP53 also serve important roles in several cancer pathways. Moreover, several small molecules target mutated TP53. Our findings indicate that TP53 signaling pathway genes carry out vital roles in WT. Nevertheless, our study has limitations, including that a model exploring other factors potentially affecting the prognosis of WT could not be constructed. Our results need to be further validated in future studies.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-6047
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6047). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Breslow N, Olshan A, Beckwith JB, et al. Epidemiology of Wilms tumor. Med Pediatr Oncol 1993;21:172-81. [Crossref] [PubMed]
- Liu J, Hua RX, Fu W, et al. MYC gene associated polymorphisms and Wilms tumor risk in Chinese children: a four-center case-control study. Ann Transl Med. 2019;7:475. [Crossref] [PubMed]
- Williams RD, Chagtai T, Alcaide-German M, et al. Multiple mechanisms of MYCN dysregulation in Wilms tumour. Oncotarget 2015;6:7232-43. [Crossref] [PubMed]
- Perlman EJ, Gadd S, Arold ST, et al. MLLT1 YEATS domain mutations in clinically distinctive Favourable Histology Wilms tumours. Nat Commun 2015;6:10013. [Crossref] [PubMed]
- Ooms AH, Gadd S, Gerhard DS, et al. Significance of TP53 Mutation in Wilms Tumors with Diffuse Anaplasia: A Report from the Children's Oncology Group. Clin Cancer Res 2016;22:5582-91. [Crossref] [PubMed]
- Yonish-Rouach E, Resnitzky D, Lotem J, et al. Wild-type p53 induces apoptosis of myeloid leukaemic cells that is inhibited by interleukin-6. Nature 1991;352:345-7. [Crossref] [PubMed]
- Lee SB, Haber DA. Wilms tumor and the WT1 gene. Exp Cell Res 2001;264:74-99. [Crossref] [PubMed]
- Kann M, Ettou S, Jung YL, et al. Genome-Wide Analysis of Wilms' Tumor 1-Controlled Gene Expression in Podocytes Reveals Key Regulatory Mechanisms. J Am Soc Nephrol 2015;26:2097-104. [Crossref] [PubMed]
- Koesters R, Ridder R, Kopp-Schneider A, et al. Mutational activation of the beta-catenin proto-oncogene is a common event in the development of Wilms' tumors. Cancer Res 1999;59:3880-2. [PubMed]
- Rivera MN, Kim WJ, Wells J, et al. An X chromosome gene, WTX, is commonly inactivated in Wilms tumor. Science 2007;315:642-5. [Crossref] [PubMed]
- Scott RH, Murray A, Baskcomb L, et al. Stratification of Wilms tumor by genetic and epigenetic analysis. Oncotarget 2012;3:327-35. [Crossref] [PubMed]
- Tang F, Min L, Seebacher NA, et al. Targeting mutant TP53 as a potential therapeutic strategy for the treatment of osteosarcoma. J Orthop Res 2019;37:789-98. [Crossref] [PubMed]
- Hunter AM, Sallman DA. Targeting TP53 Mutations in Myelodysplastic Syndromes. Hematol Oncol Clin North Am 2020;34:421-40. [Crossref] [PubMed]
- Ciccarese C, Massari F, Blanca A, et al. Tp53 and its potential therapeutic role as a target in bladder cancer. Expert Opin Ther Targets 2017;21:401-14. [Crossref] [PubMed]
- Jovanović KK, Escure G, Demonchy J, et al. Deregulation and Targeting of TP53 Pathway in Multiple Myeloma. Front Oncol 2019;8:665. [Crossref] [PubMed]
- Zandi R, Selivanova G, Christensen CL, et al. PRIMA-1Met/APR-246 induces apoptosis and tumor growth delay in small cell lung cancer expressing mutant p53. Clin Cancer Res 2011;17:2830-41. [Crossref] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Metz T, Harris AW, Adams JM. Absence of p53 allows direct immortalization of hematopoietic cells by the myc and raf oncogenes. Cell 1995;82:29-36. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007;9:166-80. [Crossref] [PubMed]
- Hsu SD, Lin FM, Wu WY, et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res 2011;39:D163-9. [Crossref] [PubMed]
- Sticht C, De La Torre C, Parveen A, et al. miRWalk: An online resource for prediction of microRNA binding sites. PLoS One 2018;13:e0206239. [Crossref] [PubMed]
- Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 2010;38:W214-20. [Crossref] [PubMed]
- Keenan AB, Torre D, Lachmann A, et al. ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res 2019;47:W212-24. [Crossref] [PubMed]
- Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [Crossref] [PubMed]
- Rees MG, Seashore-Ludlow B, Cheah JH, et al. Correlating chemical sensitivity and basal gene expression reveals mechanism of action. Nat Chem Biol 2016;12:109-16. [Crossref] [PubMed]
- Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol 2013;15:2-8. [Crossref] [PubMed]
- Goh AM, Coffill CR, Lane DP. The role of mutant p53 in human cancer. J Pathol 2011;223:116-26. [Crossref] [PubMed]
- Wu L, Xia J, Yang J, et al. Circ-ZNF609 promotes migration of colorectal cancer by inhibiting Gli1 expression via microRNA-150. J buon 2018;23:1343-9. [PubMed]
- Liu C, Yao MD, Li CP, et al. Silencing Of Circular RNA-ZNF609 Ameliorates Vascular Endothelial Dysfunction. Theranostics 2017;7:2863-77. [Crossref] [PubMed]
- Silwal-Pandit L, Russnes H, Borgen E, et al. The Sub-Cellular Localization of WRAP53 Has Prognostic Impact in Breast Cancer. PLoS One 2015;10:e0139965. [Crossref] [PubMed]
- Sohn EJ, Jung DB, Lee H, et al. CNOT2 promotes proliferation and angiogenesis via VEGF signaling in MDA-MB-231 breast cancer cells. Cancer Lett 2018;412:88-98. [Crossref] [PubMed]
- Wang Y, Zhang L, Yang J, et al. CDH13 promoter methylation regulates cisplatin resistance of non-small cell lung cancer cells. Oncol Lett 2018;16:5715-22. [Crossref] [PubMed]
- Lin XJ, He CL, Sun T, et al. hsa-miR-485-5p reverses epithelial to mesenchymal transition and promotes cisplatin-induced cell death by targeting PAK1 in oral tongue squamous cell carcinoma. Int J Mol Med 2017;40:83-9. [Crossref] [PubMed]
- Casabonne D, Benavente Y, Seifert J, et al. Serum levels of hsa-miR-16-5p, hsa-miR-29a-3p, hsa-miR-150-5p, hsa-miR-155-5p and hsa-miR-223-3p and subsequent risk of chronic lymphocytic leukemia in the EPIC study. Int J Cancer 2020;147:1315-24. [Crossref] [PubMed]
- Pan Y, Zhang R, Chen H, et al. Expression of Tripartite Motif-Containing Proteactiin 11 (TRIM11) is Associated with the Progression of Human Prostate Cancer and is Downregulated by MicroRNA-5193. Med Sci Monit 2019;25:98-106. [Crossref] [PubMed]
- Liu Y, Liu DL, Dong LL, et al. miR-612 suppresses stem cell-like property of hepatocellular carcinoma cells by modulating Sp1/Nanog signaling. Cell Death Dis 2016;7:e2377. [Crossref] [PubMed]
- Sheng L, He P, Yang X, et al. miR-612 negatively regulates colorectal cancer growth and metastasis by targeting AKT2. Cell Death Dis 2015;6:e1808. [Crossref] [PubMed]
- Wang K, Jia Z, Zou J, et al. Analysis of hsa-miR-30a-5p expression in human gliomas. Pathol Oncol Res 2013;19:405-11. [Crossref] [PubMed]
- Jin S, Liu MD, Wu H, et al. Overexpression of hsa-miR-125a-5p enhances proliferation, migration and invasion of head and neck squamous cell carcinoma cell lines by upregulating C-C chemokine receptor type 7. Oncol Lett 2018;15:9703-10. [Crossref] [PubMed]
- Osip'yants AI, Knyazev EN, Galatenko AV, et al. Changes in the Level of Circulating hsa-miR-297 and hsa-miR-19b-3p miRNA Are Associated with Generalization of Prostate Cancer. Bull Exp Biol Med 2017;162:379-82. [Crossref] [PubMed]
- Wang K, Chen M, Wu W. Analysis of microRNA (miRNA) expression profiles reveals 11 key biomarkers associated with non-small cell lung cancer. World J Surg Oncol 2017;15:175. [Crossref] [PubMed]
- Suárez-Arriaga MC, Torres J, Camorlinga-Ponce M, et al. A proposed method for the relative quantification of levels of circulating microRNAs in the plasma of gastric cancer patients. Oncol Lett 2017;13:3109-17. [Crossref] [PubMed]
- Wang H, Nie L, Wu L, et al. NR2F2 inhibits Smad7 expression and promotes TGF-β-dependent epithelial-mesenchymal transition of CRC via transactivation of miR-21. Biochem Biophys Res Commun 2017;485:181-8. [Crossref] [PubMed]
- Feng Q, Wu X, Li F, et al. miR-27b inhibits gastric cancer metastasis by targeting NR2F2. Protein Cell 2017;8:114-22. [Crossref] [PubMed]
- Wang Z, Tang F, Qi G, et al. KDM5B is overexpressed in gastric cancer and is required for gastric cancer cell proliferation and metastasis. Am J Cancer Res 2015;5:87-100. [PubMed]
- Lu PJ, Sundquist K, Baeckstrom D, et al. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J Biol Chem 1999;274:15633-45. [Crossref] [PubMed]
- Yamamoto S, Wu Z, Russnes HG, et al. JARID1B is a luminal lineage-driving oncogene in breast cancer. Cancer Cell 2014;25:762-77. [Crossref] [PubMed]
- Shen X, Zhuang Z, Zhang Y, et al. JARID1B modulates lung cancer cell proliferation and invasion by regulating p53 expression. Tumour Biol 2015;36:7133-42. [Crossref] [PubMed]
- Zhou Y, Huang T, Zhang J, et al. TEAD1/4 exerts oncogenic role and is negatively regulated by miR-4269 in gastric tumorigenesis. Oncogene 2017;36:6518-30. [Crossref] [PubMed]
- Pan D. The hippo signaling pathway in development and cancer. Dev Cell 2010;19:491-505. [Crossref] [PubMed]
- Audet-Walsh É, Vernier M, Yee T, et al. SREBF1 Activity Is Regulated by an AR/mTOR Nuclear Axis in Prostate Cancer. Mol Cancer Res 2018;16:1396-405. [Crossref] [PubMed]
- Sun M, Nie FQ, Zang C, et al. The Pseudogene DUXAP8 Promotes Non-small-cell Lung Cancer Cell Proliferation and Invasion by Epigenetically Silencing EGR1 and RHOB. Mol Ther 2017;25:739-51. [Crossref] [PubMed]
- Singh A, Trivedi P, Jain NK. Advances in siRNA delivery in cancer therapy. Artif Cells Nanomed Biotechnol 2018;46:274-83. [Crossref] [PubMed]
- Chaikomon K, Chattong S, Chaiya T, et al. Doxorubicin-conjugated dexamethasone induced MCF-7 apoptosis without entering the nucleus and able to overcome MDR-1-induced resistance. Drug Des Devel Ther 2018;12:2361-9. [Crossref] [PubMed]
- Zatloukalová P, Galoczová M, Vojtěšek B. Prima-1 and APR-246 in Cancer Therapy. Klin Onkol 2018;31:71-6. [Crossref] [PubMed]
- McKeon AM, Egan A, Chandanshive J, et al. Novel Improved Synthesis of HSP70 Inhibitor, Pifithrin-µ. In Vitro Synergy Quantification of Pifithrin-µ Combined with Pt Drugs in Prostate and Colorectal Cancer Cells. Molecules 2016;21.
- Zhou Y, Ma J, Zhang J, et al. Pifithrin-µ is efficacious against non-small cell lung cancer via inhibition of heat shock protein 70. Oncol Rep 2017;37:313-22. [Crossref] [PubMed]
- Shin D, Kim EH, Lee J, et al. RITA plus 3-MA overcomes chemoresistance of head and neck cancer cells via dual inhibition of autophagy and antioxidant systems. Redox Biol 2017;13:219-27. [Crossref] [PubMed]
- Bista M, Smithson D, Pecak A, et al. On the mechanism of action of SJ-172550 in inhibiting the interaction of MDM4 and p53. PLoS One 2012;7:e37518. [Crossref] [PubMed]
- Nenkov M, Ma Y, Haase D, et al. Growth inhibitory role of the p53 activator SCH 529074 in non-small cell lung cancer cells expressing mutant p53. Oncol Rep 2020;43:2073-82. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)