
Pentraxin-3 polymorphisms and pulmonary fungal disease in non-neutropenic patients
Introduction
Fungi are ubiquitous in the environment and approximately 400 fungal species are reported to be pathogenic to humans (1). When anatomically exposed to airborne spores, human lungs are vulnerable to certain fungal species, among which Aspergillus spp. and Cryptococcus spp. rank the top two pathogens (2). As a pattern of fungal disease, invasive fungal diseases affect more than 2 million people worldwide and cause an estimated 1.5 million deaths annually (3,4). To date, most studies have focused on life-threatening pulmonary fungal infections in immunocompromised patients. However, pulmonary fungal disease in non-neutropenic patients is not uncommon. Since 1986, a series of invasive pulmonary aspergillus (IPA) cases in non-neutropenic patients have been published (5-7). It was reported that 75.8–92.8% of the patients with Cryptococcus gattii (VGI, VGII, VGIII) infections were immunocompetent (8). A retrospective multicenter Chinese study that enrolled 474 patients clinically diagnosed with mycosis between 1998 and 2007 found that 68.7% of enrolled patients had no recognized immune dysfunction (9). Different from immunocompromised patients, non-immunocompromised patients show non-specific clinical manifestations of invasive pulmonary fungal diseases that make difficulties in distinguishing from other bacterial low respiratory infections. Azoles, amphotericin B products, and echinocandins are primary medications recommended to control fungal infections based on various conditions of patients (10). Among broad-spectrum azoles, isavuconazole is a new promising agent for both antifungal prophylaxis and treatment (11,12).
The immune response to fungal infections involves the innate immune system, which consists of physical barriers, cellular elements, and soluble factors. To contain the spread of infection, fungal pathogens are detected by families of pattern recognition receptors (PRRs) that activate and trigger the cellular and humoral immune systems (3). Long pentraxin 3 (PTX3), a PRR, plays a non-redundant role in innate immunity via pathogen recognition, agglutination, phagocytosis facilitation, and activation of complement cascade (13,14). The expression and secretion of PTX3 by neutrophils is essential for host resistance against certain microorganisms. PTX3 knockout mice show a high susceptibility to Aspergillus fumigatus in in vivo studies due to impaired conidia recognition by neutrophils, alveolar macrophages, and dendritic cells (15). Another study used a rat model in which an immunocompromised state was induced by cortisone acetate and showed that PTX3 administration cleared the respiratory tract of fungal infection and improved survival rates for lung infections (16).
In recent years, accumulated evidence suggests that single nucleotide polymorphisms (SNPs) of PTX3 are associated with a higher risk of infection by a variety of fungal diseases, including Aspergillus spp. and Candidiasis spp. (17). Most of these studies enrolled immunosuppressed individuals as objects. Cunha et al. reported that the PTX3 h2/h2 diplotype increased the risk of colonization and lung infection by species of aspergillus, thus PTX3 SNPs were regarded as a genetic predictor for aspergillosis in both stem-cell transplant recipients and lung transplant recipients (18,19). On the other hand, some underlying conditions irrelevant to the host immune status have been reported as risk factors for invasive fungal diseases. He et al. found a link between PTX3 deficiency and invasive aspergillosis in patients with chronic obstructive pulmonary disease (COPD) (20). Since COPD is considered as a “non-classic” host factor for invasive aspergillosis, it is reasonable to infer that PTX3 polymorphisms may be potentially used in patient risk stratification and help clinicians predict patient outcomes and give anti-fungal therapy guidance. To broaden our understanding, in the present study we will validate that PTX3 polymorphisms are associated with pulmonary fungal disease in non-neutropenic patients. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-5454).
Methods
The present study is a case-control study conducted in the Department of Pulmonary and Critical Care Medicine at Sun Yat-sen Memorial Hospital. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committees of Sun Yat-sen Memorial Hospital (No. SYSEC-KYKS-2020-005). Informed consent was obtained from either the patients enrolled in this study or their relatives.
Patients
Forty-five non-neutropenic Chinese patients with pulmonary fungal disease who attended the Department of Pulmonary and Critical Care Medicine at Sun Yat-sen Memorial Hospital between January 2018 and December 2019 were included in this study; these patients comprised the case group. Of these patients, 25 had pulmonary aspergillosis, 18 had pulmonary cryptococcosis, and 2 had other types of pulmonary fungal disease (1 case of Talaromyces marneffei and 1 case of Candida albicans). All cases were diagnosed by two senior respiratory physicians (Tang T and Huang L) based on clinical features and anti-fungal treatment response.
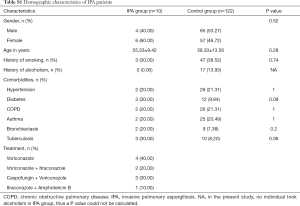
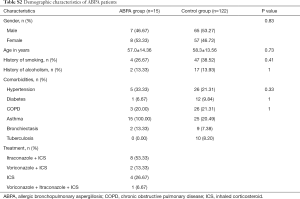
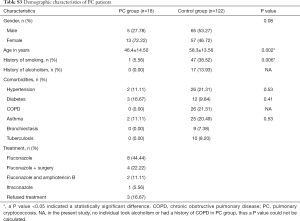
Of the patients with aspergillosis, 10 were diagnosed with “proven” (n=4) and “probable” (n=6) IPA according to the criteria of the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) (10) and 15 were diagnosed with allergic bronchopulmonary aspergillosis (ABPA) according to the criteria of the International Society for Human and Animal Mycology (ISHAM) (21). The 4 “proven” IPA patients were pathologically confirmed by transbronchoscopic lung biopsy (TBLB) (n=3) or computed tomography (CT)-guided percutaneous transthoracic biopsy (n=1). Of the “probable” IPA patients, 4 had two positive sputum cultures for Aspergillus spp. and the remainder showed positive results in Galactomannan testing of bronchoalveolar lavage fluid. All “probable” IPA cases showed a good clinical response to anti-fungal treatment (voriconazole or itraconazole), which confirmed the pulmonary aspergillosis diagnoses. Of the patients with cryptococcosis, all were diagnosed according to the criteria of the Infectious Diseases Society of America (IDSA) (22). Among them, 11 were diagnosed by pathological examination (1 case by TBLB, 6 cases by CT-guided transthoracic biopsy, and 4 cases by surgical biopsy), and 7 were diagnosed by a positive serum cryptococcal capsular polysaccharide antigen test. More clinical features of the patients are listed below in the Supplementary file (Tables S1-S3).
Full table
Full table
Full table
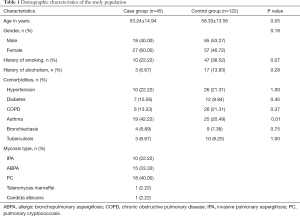
We obtained the medical records of patients without mycosis that presented to the same department during the same time period (from January 2018 to December 2019) and randomly selected 122 as the control group (see Supplementary file “Calculation of sample size”). The key inclusion criteria were that patients were older than 18 years. Exclusion criteria for both the case group and the control group included the following: a history of neutropenia (defined as neutrophil count <1.5×109/L), solid organ transplantation (SOT), hematopoietic stem-cell transplantation (HSCT), or malignancies. The demographic characteristics of the study population are showed in Table 1.
Full table
Genotyping strategy and DNA extraction
In line with previous studies (18-20,23), three SNPs (rs2305619, rs3816527, and rs1840680) of PTX3 were selected. The genotypes are shown in Table 2.

Full table
Peripheral blood obtained from each subject was isolated by erythrocyte lysate (Sango Biotech, Shanghai, China) according to the manufacturer’s instructions. Genomic DNA was extracted using a genomic DNA extraction kit (Shanghai Generay Biotech, China).
Real-time polymerase chain reaction
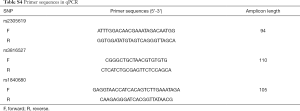
The process of real-time polymerase chain reaction (PCR) was performed in triplicate using the TaKaRa Premix Ex Taq II kits (TaKaRa Bio Inc.) according to the manufacturer’s protocol. The primers and minor groove binder (MGB) probes (Sango Biotech, Shanghai, China) were designed in Beacon Designer software based on the SNP sequences located in the National Center for Biotechnology Information (NCBI) SNP database. The primers used in this study are listed in Table S4.
Full table
Defining SNP genotypes
Using fluorescent probes, melting curve analyses were used to define the SNP genotypes, which were analyzed by CFX Manager Software 1.6 (Bio-Rad).
Statistical analysis
Quantitative data were analyzed by the Wilcoxon rank-sum test or the Student’s t-test; data were expressed as mean ± standard deviation. Categorical data were analyzed by the chi-squared test or Fisher’s exact test; data were expressed as n (percentage). Logistic regression was used to analyze the associations between SNPs and different pulmonary fungal diseases. These correlations were expressed as odds ratios (ORs) and 95% confidence intervals (CIs). A P value <0.05 indicated a statistically significant difference. All data were analyzed by R-studio 3.5. Haplotype analyses and HWE were used to describe the PTX3 SNP genotypes. These analyses were carried out using HaploView version 4.2 statistical software.
Results
Patient characteristics
The patients in the two groups were age and gender matched. There were no statistically significant differences between the case group and the control group in terms of smoking history, alcoholism, or comorbidities (Table 1).
Hardy-Weinberg equilibrium analysis
The three SNPs (rs2305619, rs3816527, and rs1840680) were found to be in accordance with Hardy-Weinberg equilibrium (HWE) in the whole study population. The minor allele frequencies (MAFs) were large than 5%. Both the case group and the control group represented population genetic stability in the context of P value >0.05 (Table 2).
Linkage disequilibrium analysis of SNPs
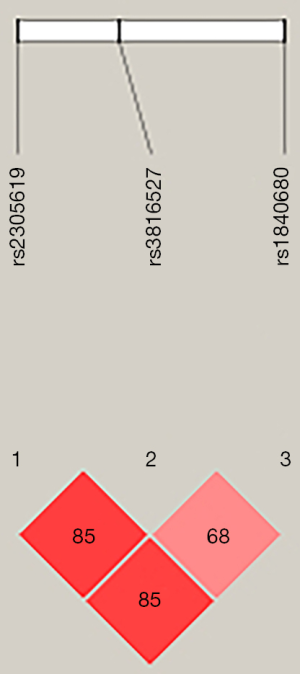
Paired linkage disequilibrium was calculated between the three SNPs. The analysis showed that rs2305619 was in linkage disequilibrium with rs3816527 (D’=0.85; Figure 1). Similarly, linkage disequilibrium was observed between rs2305619 and rs1840680 (D’=0.85; Figure 1).
SNP associations with pulmonary fungal disease
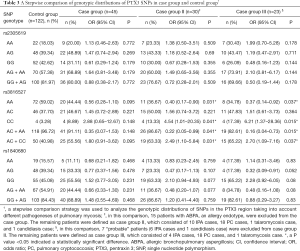
We first analyzed the genetic distribution in the study population of SNPs in the PTX3 region. No SNP genotypic distribution differences were observed between the case group and the control group (Table 3). At the same time, there were no differences in SNP haplotype frequencies found between the two groups (Table 4).
Full table

Full table
We then performed a stepwise analysis to find out whether mycosis type had an impact on SNP distribution and frequency. ABPA is an allergic form of aspergillosis, whereas IPA, cryptococcosis, talaromycosis, and pulmonary candidiasis are invasive fungal diseases. Taking this into consideration, we excluded ABPA patients from the case group and analyzed the remaining case group patients; this was case group II. We identified a significant difference in SNP rs3816527 distribution between case group II and the control group. This suggested that SNP rs3816527 CC homozygosity might be associated with an increased risk for invasive mycoses (OR, 4.54; 95% CI, 1.01–20.35; P=0.041), while AA homozygosity might be associated with a lower risk (OR, 0.40; 95% CI, 0.17–0.90; P=0.031). No genotypic distribution differences for the other two SNPs (rs2305619 and rs1840680) were observed between case group II and the control group (Table 3).
To further confirm the association between SNPs and invasive pulmonary fungal disease, we excluded the 7 “probable” cases from case group II (6 IPA cases and 1 candidiasis case) and analyzed the remaining “proven” cases as case group III. Similarly, a significant difference in SNP rs3816527 distribution was found between case group III and the control group. In term of haplotype analysis, we found that rs3816527 CC homozygosity was associated with an increased risk for “proven” mycoses (OR, 6.21; 95% CI, 1.37–28.36; P=0.015), whereas AA homozygosity was associated with a decreased risk (OR, 0.37; 95% CI, 0.14–0.92; P=0.037). No genotypic differences were observed for the other two SNPs (rs2305619 and rs1840680) (Table 3).
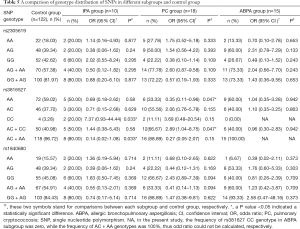
Finally, we performed a subgroup analysis to determine the mycosis types with which SNPs are associated. Consistent with our previous results, SNP rs3816527 CC homozygosity was associated with an increased risk for mycoses, but this result was only found for the IPA subgroup (OR, 7.37; 95% CI, 0.93–44.44; P=0.033). SNP rs3816527 AA homozygosity was found to be associated with a lower risk for mycoses, but this only impacted the cryptococcus subgroup (OR, 0.35; 95% CI, 0.11–0.96; P=0.047). There were no significant genotype differences found between the ABPA group and the control group, and no genotypic distribution differences were observed for the other two SNPs (rs2305619 and rs1840680) (Table 5).
Full table
Discussion
In the present case-control study of non-neutropenic patients, we demonstrated that one PTX3 SNP, rs3816527, was associated with invasive pulmonary mycosis. Although the haplotypes of the three SNPs did not reveal any statistical differences between patients with pulmonary fungal disease and control patients, we still found that the rs3816527 CC homozygote was associated with a higher risk of IPA and the rs3816527 AA homozygote was associated with a lower risk of pulmonary cryptococcus.
There are several clinical conditions that enhance the risk of infection by invasive mold, including HSCT, SOT, hematological cancer, acquired immune deficiency syndrome (AIDS), long-term corticosteroids or other immunosuppressive therapies, et cetera. Several well-conducted studies have demonstrated that patient PTX3 genetic polymorphisms may influence their susceptibility to fungal pathogens. A study by Cunha et al. (18) highlighted the potential role of donor homozygous haplotype h2/h2 (rs2305619-rs3816527: G-A/G-A) in the development of invasive aspergillosis in HSCT recipients. This result was replicated in another observational cohort study that enrolled 2,609 HSCT cases (24). Further, it has been reported that PTX3 rs2305619 and/or rs3816527 homozygosity is an independent predictor for invasive mold infections in acute leukemia patients undergoing intensive chemotherapy that did not have pre-existing neutropenia (HR =5.06; 95% CI, 1.68–15.2, P=0.004) (25). Similarly, an association between the PTX3 SNP rs3816527 and mycoses in SOT recipients has been established (23). After adjusting for all relevant covariates, the authors found that the rs3816527 AA genotype greatly influenced susceptibility to fungal colonization and infection. The presence of PTX3 SNPs, notably rs3816527, in different immunodeficiency patient populations suggests that PTX3 SNPs may increase these patients’ risk for fungal infections.
Our findings are not entirely consistent with earlier researches; this may be due to population heterogeneity. In this single-center study, all the patients were from the South China. Another study of COPD patients from Southeast China (20) did not observe an association between the rs3816527 AA homozygote and IPA; this was similar to our results. Contrary to expectations, however, only the SNP rs1840680 AA homozygote was found to be associated with increased susceptibility to IPA; this had not been previously reported. Another possible reason for the discrepancies between the present study and previous studies could be the small sample size of subjects with the SNP rs3816527 CC homozygote. There are significant regional differences in rs3816527 C allele frequency. According to the Single Nucleotide Polymorphism database (26), rs3816527 C allele frequency in the European population is 0.398, while in China and Africa it is only 0.243–0.256. A multi-racial investigation focused on non-neutropenic patients is therefore needed.
The PTX3 gene is located in a phylogenetically conserved region. Genetic variants may therefore affect messenger RNA stability and PTX3 protein production, which influences susceptibility to fungal infections (18). It is of note that the rs3816527 AA homozygote seemed to be related to a lower risk for pulmonary cryptococcosis in the present study. Cryptococcus species produce a mucoid capsule that surrounds the yeast cells during infection. As the outermost and most abundant component of the capsule, glucuronoxylomannan (GXM) not only interferes with host recognition of β-glucans and mannoproteins within the cell wall (27), but also directly interacts with FcγRIIB, producing inhibitory signals that contribute to immune tolerance (28). To date, several PRRs involved in the recognition of GXM have been identified, including C-type lectin receptors (CLRs), Toll-like receptors (TLRs), and NOD-like receptors (NLRs) (29). Different from PTX3, theses receptors are cell-associated instead of soluble. Although a PTX3 genetic variation was found to be associated with a reduced risk for cryptococcosis, further investigation is needed to validate the potential protection effects of PTX3.
There was no difference in genotypic distribution for PTX3 SNPs found between the ABPA subgroup and the control group. Once inhaled into the respiratory tract, aspergillus conidia colonize and germinate, causing type I hypersensitivity instead of tissue invasion (30). In resting conidia, β (1,3)-glucans in the cell wall are concealed by a layer of hydrophobic proteins called rodlets that are shed during germination (23). As a result, PTX3 is prevented from recognizing its binding sites. Garlanda et al. also found that PTX3 in human endothelial cells and epithelial cells was produced in little amounts with aspergillus conidia stimulation (15). On the other hand, polymorphisms in mannose-binding lectin and surfactant proteins have been observed in several small cohorts, which were considered contributing to ABPA (31). These findings may support the hypothesis that PTX3 polymorphisms are not involved in the pathogenesis of ABPA. As is known, ABPA belongs to the aspergillosis clinical spectrum and its initial treatment includes oral corticosteroids (32). Our findings indicate that patients with ABPA may have a low risk of developing invasive aspergillosis in spite of corticosteroid medications.
Several limitations must be noted with regards to the present study. First, this was a single-center study in Southeast China with limited ethnic diversity; this may limit the extent these results can be generalized. Second, as a retrospective study, we did not measure patient PTX3 mRNA levels or protein levels at the onset of their pulmonary fungal disease. Functional studies are therefore required to determine the effect of different SNPs on the risk of developing different pulmonary fungal diseases.
Conclusions
We found that the SNP rs3816527 was associated with pulmonary fungal disease in non-neutropenic patients; the rs3816527 CC homozygote was related to an increased risk of IPA. Identifying PTX3 genetic variations associated with pulmonary fungal disease may be beneficial for the risk stratification of non-neutropenic patients, as well as classical immunocompromised patients. Further investigations are needed to validate this genetic predisposition to pulmonary fungal disease in larger populations. Functional studies are also required.
Supplementary
Calculation of sample size
We recruited 45 patients with pulmonary fungal disease as the case group (N1). The sample size of control group (N2) was calculated using the following formula:
, (N2=cN1, , β=0.1, α=0.05)
Note: P1 is the SNP frequency in patients with pulmonary fungal disease; P2 is the SNP frequency in patients without pulmonary fungal disease.
In accordance with a previous study (17), we chose rs1840680, which had a P1 value of 0.417 and a P2 value of 0.117. According to the calculation, the control group sample size (N2) had to be no less than 72 patients. In order to maintain HWE, we enrolled 122 of 766 inpatients as a control group using systematic sampling.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (81670022); the Natural Science Foundation of Guangdong Province (2017A030313681); and the Guangzhou Municipal Science and Technology Project (201704020123).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-5454
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-5454
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-5454). The authors report grants from the National Natural Science Foundation of China, grants from the Natural Science Foundation of Guangdong Province, grants from the Guangzhou Municipal Science and Technology Project, during the conduct of the study.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committees of Sun Yat-sen Memorial Hospital (SYSEC-KY-KS-2020-005). Informed consent was obtained from either the patients enrolled in this study or their relatives.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Taylor LH, Latham SM, Woolhouse ME. Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci 2001;356:983-9. [Crossref] [PubMed]
- Enoch DA, Yang H, Aliyu SH, et al. The changing epidemiology of invasive fungal infections. Methods Mol Biol 2017;1508:17-65. [PubMed]
- Kumar V, van de Veerdonk FL, Netea MG. Antifungal immune responses: emerging host-pathogen interactions and translational implications. Genome Med 2018;10:39. [PubMed]
- Underhill DM, Pearlman E. Immune interactions with pathogenic and commensal fungi: A Two-Way Street. Immunity 2015;43:845-58. [Crossref] [PubMed]
- Karam GH, Griffin FM Jr. Invasive pulmonary aspergillosis in nonimmunocompromised, nonneutropenic hosts. Rev Infect Dis 1986;8:357-63. [Crossref] [PubMed]
- Clancy CJ, Nguyen MH. Acute community-acquired pneumonia due to Aspergillus in presumably immunocompetent hosts: clues for recognition of a rare but fatal disease. Chest 1998;114:629-34. [Crossref] [PubMed]
- Ko JP, Kim DH, Shepard JA. Pulmonary aspergillosis in an immunocompetent patient. J Thorac Imaging 2002;17:70-3. [Crossref] [PubMed]
- Chen SC, Meyer W, Sorrell TC. Cryptococcus gattii infections. Clin Microbiol Rev 2014;27:980-1024. [Crossref] [PubMed]
- Liu YN, She DY, Sun TY, et al. A multicenter retrospective study of pulmonary mycosis clinically proven from 1998 to 2007. Zhonghua Jie He He Hu Xi Za Zhi 2011;34:86-90. [PubMed]
- De Pauw B, Walsh TJ, Donnelly JP, et al. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis 2008;46:1813-21. [Crossref] [PubMed]
- Maertens J, Selleslag D, Heinz WJ, et al. Treatment outcomes in patients with proven/probable vs possible invasive mould disease in a phase III trial comparing isavuconazole vs voriconazole. Mycoses 2018;61:868-76. [Crossref] [PubMed]
- Samanta P, Clancy CJ, Marini RV, et al. Isavuconazole is as effective as and better tolerated than voriconazole for antifungal prophylaxis in lung transplant recipients. Clin Infect Dis. 2020 May 28:ciaa652. Epub ahead of print.
- Mantovani A, Valentino S, Gentile S, et al. The long pentraxin PTX3: a paradigm for humoral pattern recognition molecules. Ann N Y Acad Sci 2013;1285:1-14. [Crossref] [PubMed]
- Daigo K, Mantovani A, Bottazzi B. The yin-yang of long pentraxin PTX3 in inflammation and immunity. Immunol Lett 2014;161:38-43. [Crossref] [PubMed]
- Garlanda C, Hirsch E, Bozza S, et al. Non-redundant role of the long pentraxin PTX3 in anti-fungal innate immune response. Nature 2002;420:182-6. [Crossref] [PubMed]
- Lo Giudice P, Campo S, Verdoliva A, et al. Efficacy of PTX3 in a rat model of invasive aspergillosis. Antimicrob Agents Chemother 2010;54:4513-5. [Crossref] [PubMed]
- Wójtowicz A, Bochud PY. Host genetics of invasive Aspergillus and Candida infections. Semin Immunopathol 2015;37:173-86. [Crossref] [PubMed]
- Cunha C, Aversa F, Lacerda JF, et al. Genetic PTX3 deficiency and aspergillosis in stem-cell transplantation. N Engl J Med 2014;370:421-32. [Crossref] [PubMed]
- Cunha C, Monteiro AA, Oliveira-Coelho A, et al. PTX3-based genetic testing for risk of aspergillosis after lung transplant. Clin Infect Dis 2015;61:1893-4. [Crossref] [PubMed]
- He Q, Li H, Rui Y, et al. Pentraxin 3 Gene Polymorphisms and Pulmonary Aspergillosis in Chronic Obstructive Pulmonary Disease Patients. Clin Infect Dis 2018;66:261-7. [Crossref] [PubMed]
- Agarwal R, Chakrabarti A, Shah A, et al. Allergic bronchopulmonary aspergillosis: review of literature and proposal of new diagnostic and classification criteria. Clin Exp Allergy 2013;43:850-73. [Crossref] [PubMed]
- Perfect JR, Dismukes WE, Dromer F, et al. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the infectious diseases society of america. Clin Infect Dis 2010;50:291-22. [Crossref] [PubMed]
- Wójtowicz A, Lecompte TD, Bibert S, et al. PTX3 Polymorphisms and Invasive Mold Infections After Solid Organ Transplant. Clin Infect Dis 2015;61:619-22. [Crossref] [PubMed]
- Fisher CE, Hohl TM, Fan W, et al. Validation of single nucleotide polymorphisms in invasive aspergillosis following hematopoietic cell transplantation. Blood 2017;129:2693-701. [Crossref] [PubMed]
- Brunel AS, Wójtowicz A, Lamoth F, et al. Pentraxin-3 polymorphisms and invasive mold infections in acute leukemia patients receiving intensive chemotherapy. Haematologica 2018;103:e527-e530. [Crossref] [PubMed]
- Single Nucleotide Polymorphism Database (dbSNP). Available online: ; latest visit on July 10th, 2020.https://www.ncbi.nlm.nih.gov/snp
- Zaragoza O, Rodrigues ML, De Jesus M, et al. The capsule of the fungal pathogen Cryptococcus neoformans. Adv Appl Microbiol 2009;68:133-216. [Crossref] [PubMed]
- Piccioni M, Monari C, Bevilacqua S, et al. A critical role for FcgammaRIIB in up-regulation of Fas ligand induced by a microbial polysaccharide. Clin Exp Immunol 2011;165:190-201. [Crossref] [PubMed]
- Patin EC, Thompson A, Orr SJ. Pattern recognition receptors in fungal immunity. Semin Cell Dev Biol 2019;89:24-33. [Crossref] [PubMed]
- Feng H, Lv P, Ren X, et al. Misinterpretation of allergic bronchopulmonary aspergillosis/allergic bronchopulmonary mycosis due to diverse characteristics in different clinical stages. J Thorac Dis 2019;11:4484-91. [Crossref] [PubMed]
- Ok M, Einsele H, Loeffler J. Genetic susceptibility to Aspergillus fumigatus infections. Int J Med Microbiol 2011;301:445-52. [Crossref] [PubMed]
- Greenberger PA, Bush RK, Demain JG, et al. Allergic bronchopulmonary aspergillosis. J Allergy Clin Immunol Pract 2014;2:703-8. [Crossref] [PubMed]
(English Language Editor: B. Madden)


