A better method for the dynamic, precise estimating of blood/haemoglobin loss based on deep learning of artificial intelligence
Introduction
Intra-operative blood loss can be rapid and extensive, which affects the transfusion decisions of perioperative management. One of the most important responsibilities of an anesthesiologist is to estimate intra-operative blood loss in a timely and accurate manner and make decisions about invasive monitoring and blood transfusion. The underestimation of blood loss may result in a haemoglobin (Hb) deficiency resulting from a delayed transfusion, which leads to circulation instability and an insufficient oxygen supply. Furthermore, the overestimation of blood loss may be associated with increased morbidity and possible mortality from unnecessary invasive monitoring techniques, as well as a risk of wastage of blood products (1). There are several methods or technologies used to estimate blood loss (EBL) or Hb loss (EHL) during surgery (2), such as the visual method, gravimetric method and, recently, new methods based on computer algorithms (3-6). However, these methods present obvious limitations. Despite its accuracy and convenience in clinical use, the gravimetric method is not recommended. Although most frequently used in routine work, visual estimation by medical staff is not very reliable or accurate, especially in surgical cases with massive bleeding (2). Furthermore, the accuracy of visual EBL appears to be independent of sex, training and experience (7), which indicates that the method itself is inaccurate and unreliable.
Recently, a new method (Triton System, Gauss Surgical, Inc., Los Altos, USA) based on artificial intelligence (AI) has been introduced in some hospitals in the United States of America (3-6). Triton System is a camera-enabled mobile application native to the iPad and is mainly used for EHL. During surgery, captured images of blood-soaked sponges are encrypted, transferred wirelessly to a remote server and analysed by AI methods; then, the real-time EBL and EHL will present data based on the operation start time (6). Although this approach seemed promising in clinical situations [the bias and limits of agreement (LOA) were within the clinically relevant differences] (4,6), this system still has significant shortcomings and needs to be improved. First, the accuracy of this system needs to be further increased by new AI technology. According to the patent published in 2013 (8), the key technology of the image processing in Triton System is feature extraction technology (FET), which in brief, extracts the features of images, such as intensity, luminosity, hue, and other colour-related values, for EHL. However, the algorithm for EHL is unknown. Referring to the published data, linear regression (LR) is the most likely algorithm used for EHL. LR is used to determine the quantitative relationship between two or more variables and is one of the most commonly used statistical algorithms for continuous data. The major limitation of LR is that it does not work well when the realistic relationships among variables are non-linear. The relationship between features extracted by FET and Hb loss may be non-linear and more complex. Triton System has indicated that feature extraction may be sufficient for processing images of blood-soaked sponges; however, new algorithms such as random forest (RF) (9) and extreme gradient boosting (Xgboost) (10) may increase the accuracy. Recently, as another branch of AI, deep learning has increasingly been used in the medical field and has the advantage of self-learning, resulting in automatic extraction of features. Densely connected convolutional networks (DenseNet) are one of the most promising deep learning algorithms (11), which can further increase the accuracy of image recognition and segmentation (12,13). These advances indicate that applying new AI technologies, including modified algorithms and deep learning, may increase accuracy and decrease bias. Second, Triton System is mainly used for EHL, and EBL is calculated by EHL (EBL = EHL/preoperative concentration of Hb). In clinical situations, the volume of blood loss is more important for medical staff due to the demand of guiding intra-operative fluid management. The EBL determined by Triton is not as acceptable because Hb changes in the surgical process. Third, the actual Hb in the sponge is detected by a low-concentration Hb analyser after rinsing (6). Although researchers adjusted the value of Hb, they could not eliminate the systemic biases of rinsing and the analyser (5). Applying a different AI technology and alternative blood-soaked sponge models with a known volume and Hb concentration with non-recycled blood may increase the accuracy of the estimation. Therefore, we aimed to provide a better method based on DenseNet to estimate blood loss dynamically and directly and Hb loss.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-1806).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was approved by the Institutional Ethics Committee of the First Affiliated Hospital of Third Military Medical University (No. KY2019127), and written informed consent was obtained from each patient. The principal researcher was Prof. Bin Yi. Herein, we developed estimating models based on different AI technologies using images of blood-soaked sponges. The blood-soaked sponges contained surgical patients’ non-recycled blood and were collected from a suction canister. Blood collection and image acquisition were completed at the First Affiliated Hospital of Third Military Medical University in Chongqing, China, between October 30, 2019, and November 15, 2019.
Blood collection, blood-soaked sponge preparation and image acquisition
The inclusion criteria were as follows: patients were willing to participate in the study and signed the informed consent form; the intra-operative blood loss was greater than 100 mL, and the blood Hb concentration in the suction canister was above 60 g/L. The exclusion criteria were as follows: patients refused to participate; patients were scheduled for caesarean section; patients were with pleural effusion and ascites, or with diseases which would change the colour of the blood, such as pancreatitis, hypoxia, jaundice, carbon monoxide poisoning, nitrite poisoning, and so on, or with blood-borne diseases, such as hepatitis B, hepatitis C, AIDS and so on.
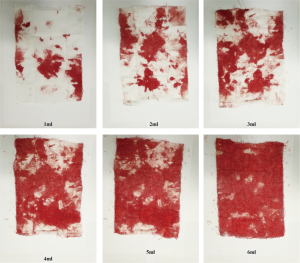
On the surgical day, the suction canister was pretreated with 600 U/mL heparin sodium with a volume of 5 mL. Before the abundant flushing of fluid intra-operatively, researchers collected 20 mL of non-recycled blood in a syringe and performed blood gas analysis to detect the concentration of Hb. Preparation of the blood-soaked sponge was carried out within two hours after collection. The surgical sponges were 6 cm × 8 cm. In our preliminary experiment with artificial blood, 7 mL artificial blood was sufficient to soak the sponge with blood. In the current study, we set the gradient of blood as follows: 1, 2, 3, 4, 5, and 6 mL (shown in Figure S1). There were four researchers, two for the blood-soaked model preparation and the other two for image acquisition. One researcher injected a set volume of the blood sample into a bowl, and the other researcher wiped the blood with a sponge. Then, the blood-soaked sponge was fully expanded and placed on white paper. The two researchers exchanged roles every five samples during the whole process. To simulate the clinical use of sponges and to make the most of the non-recycled blood, we added 1 mL of non-recycled blood to the bowl each time until 6 ml had been added. Therefore, 20 mL non-recycled blood could be used to establish three different sets of blood-soaked images (18 images in total). In the image acquisition step, the camera was adjusted to the same height and captured images with the same parameters. One researcher took images of the blood-soaked sponges in the normal illumination for the operation room, while the other recorded information about the images.
Establish models based on feature engineering method
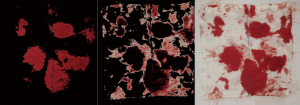
As shown in Figure 1, the process of establishing models based on the feature engineering method comprised three steps, namely, image preprocessing, feature extraction and estimation. Finally, we collected non-recycled blood form 34 surgical patients. Therefore, 569 portions of blood-soaked images were employed for feature engineering. After image acquisition, the first step of image preprocessing was to resize the image to 480×480×3. For feature engineering, the blood area was extracted. There were two steps for the blood area extraction. First, the resized images were converted from the RGB colour space into the HSV colour space. The value of a pixel in the H channel ranged from 0 to 180, and the values pixel in the S channel and V channel ranged from 0 to 255. Second, two mask maps were established to generate two blood area images that were composed of two colours, namely, black and red (shown in Figure S2); the red area represented the blood area. The detailed information is shown in the supplementary materials.
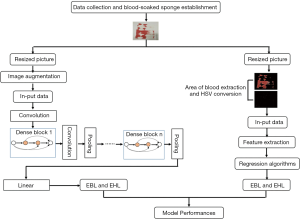
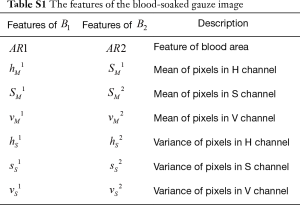
As is known, the area size, colour depth and brightness are associated with blood loss and Hb loss. After image preprocessing, feature extraction began. The two blood area images only had two colours. As a result, the two area features could be represented by the proportion of pixels in the blood area to the total pixels in the H channel of the two blood area images. The area size of the two blood images was calculated by the number of pixels. In the HSV colour space, the three channels represented hue, saturation and brightness. Therefore, as for colour depth and brightness, the mean and variance of the H, S and V channels were used to represent the colour depth and brightness of the blood area. After image preprocessing, there were 14 features of the blood-soaked sponge images, namely, the area sizes of the blood and the mean and variance of the H, S and V channels in the two blood-area images (shown in Table S1). After image preprocessing, the dataset was randomly divided into training dataset and testing dataset. In the current study, 10-flod cross validation was used to form the training and testing datasets for methods based on the feature engineering and deep learning. First of all, we arranged the samples in ascending order according to the gradients of blood loss. Then, we randomly shuffled the order of the samples and divided them into 10 equal subsets, in which contained all the gradients blood loss. While the proportion among the number of samples corresponding to the gradients of blood loss was slightly different in each subset. Afterwards, each subset was then used once as a testing dataset while the 9 remaining subsets formed the training dataset. As a consequence, the ratio of the number of samples between the training dataset and the testing dataset was about 9:1.
Full table
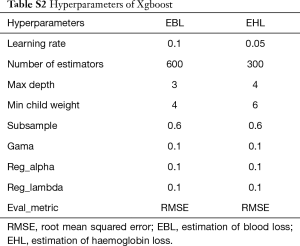
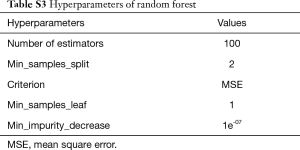
In the current study, we chose three different kinds of models that are widely used in machine learning. After feature extraction, the models for EBL and EHL were based on regression algorithms, namely, Xgboost, LR, and RF. LR is a linear model that uses a linear combination of attributes to make predictions with a good interpretability. RF can be used for category and regression, which are fit for high-dimensional data. Xgboost is an integrated learning method proposed by Chen et al. (14) with an excellent learning effect and efficient training speed, which have been widely accepted. The hyperparameters are fine-tuned through training dataset. And the detailed information about hyperparameters of Xgboost and RF for EBL and EHL were presented in the supplementary materials (shown in Tables S2 and S3).
Full table
Full table
Establish models based on dense network method
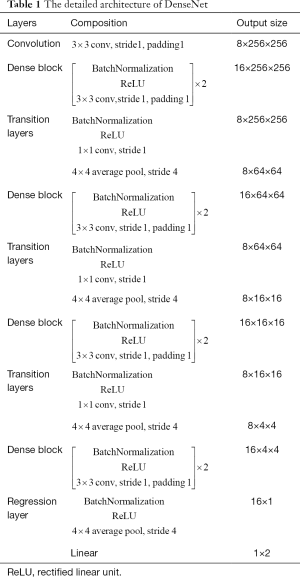
Images were divided into training and testing datasets as previously described. Since more images would be required for deep learning, we did image augmentation. In the training process, an online augmentation method was used to increase the number of the samples, which employed horizontal flip, vertical flip and horizontal vertical flip to generate new samples. The number of samples was quadrupled by this way. At each batch, the augmentation method was applied on the samples before training. The experimental environment of this article was based on a GPU supercomputing cluster server consisting of three FitServer R4200s. Ubuntu 16.04 LTS was used as the operating system with an Intel Xeon e5-2620 V4 processor and Nvidia GTX 1080 Ti GPU; the memory was 128 GB (shown in Figure S3). Pytoch was used to build the convolutional neural network, and Python 3.6 was used as the programming language. DenseNet was proposed by Huang et al. in 2016 (11), and similar to previous deep learning algorithms, the resized images could be directly entered into the model for automatic learning. A feedforward approach was used to connect each layer to each of the other layers. For each layer in this network, the feature maps of all previous layers were used as inputs, while its own feature map was used as the input for all subsequent layers. To further improve the accuracy of EBL and EHL, DenseNet was applied in the current study. To reduce the number of parameters in the DenseNet, we did some modifications in DenseNet in this paper. In detail, the input in the presented DenseNet consisted one 3×3 convolutional layer. Second, the kernel size of average pooling layers in the presented DenseNet was 4×4.
As shown in Figure 1, first, we resized the image as 256×256×3. At the beginning, a 3×3 convolution with 8 output channels was performed on the input images. Then, there were four dense blocks and three transitional layers in DenseNet. The dense block contains two layers. Each layer can be thought of as a composite function of three consecutive operations: bath normalization, followed by a rectified linear unit and a 3×3 convolution. For the 3×3 convolutional layers in this network, each side of the inputs was zero-padded by one pixel to keep the feature map size fixed. The transition layers consist of batch normalization (BN), ReLU, 1×1 convolution and 4×4 average pooling. At the end of the last dense block, the architecture consisted of a bath normalization and rectified linear unit followed by a 4×4 average pooling layer, and then, a linear layer was attached to obtain the output. The output is 1D vector that consists of two values which are the predictive values of EBL and EHL (shown in Table 1).
Full table
The MSE loss function was used within training processes of DenseNet. n is the number of the samples,
[1]
The growth rate of DenseNet was 4. The optimizer is Adam and the compression factor of DenseNet is 0.5. The training of DenseNet is controlled by the epoch without stopping criteria. The maximum epoch is 150. When the epoch is below 50, the learning rate is 0.005; when the epoch is between 50 to 100, the learning rate is 0.0005; When the epoch is between 100 and 150, the learning rate is 0.00005. The training Batch size is 20, while the testing batch size 8.
Model evaluation
EBL and EHL were estimated from continuous data, so we evaluated the performance of the models with regression indicators, such as the mean absolute error (MAE), mean square error (MSE) and R-squared (R2). The MAE is used to describe the average of difference between the predicted value and the actual value. The MSE represents the average of the square of the difference between the estimated value and the actual value. Usually, the MSE is more sensitive than the MAE. For the MAE and MSE, the smaller the value, the better. R2, also called the coefficient of determination, the closer the value to 1, the stronger the ability to interpret the output and the better the model fits.
Statistical analysis
For quantitative variables, the mean, standard deviation (SD), and range are presented. For the primary effectiveness variables, 95% confidence intervals (CIs) are presented. The concordance between methods based on AI and the actual value was tested via a Bland-Altman analysis, wherein the bias (the mean difference between the two measures), upper or lower LOA with 95% CIs and standard deviation of error were calculated by MedCalc Statistical Software version 15.8 (MedCalc Software bvba, Ostend, Belgium; https://www.medcalc.org; 2015).
Results
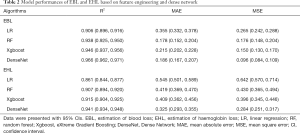
As shown in Table 2, for EBL, the R2 value was greater than 0.900, which was quite satisfactory regardless of the algorithm. However, taking the MSE and MAE into account, the methods based on RF and Xgboost seemed better than those based on LR. The R2, MAE and MSE values for the method based on DenseNet were 0.966 (95% CI: 0.962–0.971), 0.186 (95% CI: 0.167–0.207) and 0.096 (95% CI: 0.084–0.109), respectively, which were more satisfactory than those of the methods based on feature engineering. For EHL, the estimation was more difficult. Among the methods based on feature engineering, Xgboost presented a higher R2 values (0.915, 95% CI: 0.904–0.925) and lower MAE (0.409, 95% CI: 0.362–0.456) and MSE values (0.396, 95% CI: 0.345–0.446). However, the R2, MAE and MSE values for the method based on DenseNet were 0.941 (95% CI: 0.934–0.948), 0.325 (95% CI: 0.293–0.355) and 0.284 (95% CI: 0.251–0.317), respectively, which were more satisfactory than those of Xgboost.
Full table
As shown in Table 3 and Figure 2, we applied the Bland-Altman analysis to evaluate the concordance of EBL and EHL among the different methods based on AI and the actual blood and Hb losses. For EBL, the biases of Xgboost (−0.04 mL) and DenseNet (0.02 mL) were the smallest. In addition, the LOA of DenseNet (−0.47 mL to 0.52 mL) were the narrowest. For EHL, the biases of the methods based on Xgboost (−0.05 g) and DenseNet (0.05 g) were the smallest. In addition, the LOA based on DenseNet (−0.87 g to 0.97 g) were the narrowest (shown in Table 3 and Figure 3). In our study, not only the model performance but also the results of the concordance analysis for the method based on LR were not superior to those of other methods based on AI.
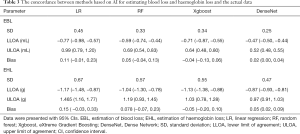
Full table
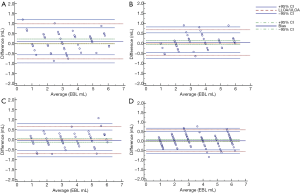
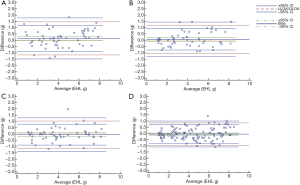
Discussion
In the current study, we developed better methods for EBL and EHL based on feature engineering and deep learning using blood-soaked sponges. For EBL and EHL, regardless of comparing the model performance or concordance results, the methods based on DenseNet were more appropriate than the model based on LR.
The precise and dynamic estimation of intra-operative blood loss is one of the medical staff’s key jobs and is also quite important for surgical patients’ safety. The aim of EBL and EHL is to guide perioperative fluid management and surgical decisions. Therefore, most of the estimation methods are for EBL, not for EHL. Until now, visual estimation has been the most frequently used method. However, as we described previously, visual estimation is inaccurate and unreliable; in particular, the larger the amount of blood loss, the larger the measurement error (2). Moreover, other studies have suggested that overestimations and underestimations have no obvious association with the experience of medical staffs (15,16). Although some visual aid tools and training courses have been reported to increase the accuracy of visual EBL (17-20), the visual aids were for a particular situation or for a set amount of blood loss, which not be very practical or helpful for the complicated, variable intra-operative requirements for EBL. The gravimetric method involves the collection of all blood-soaked items and deduction of the dry weight of the item, which requires an accurate scale. However, this method cannot discriminate between blood and other types of fluid, which may affect the final results. Furthermore, although this method is easy to master, it is time-consuming and labor-intensive (2). Photometric methods require specific devices, which are not practical for daily use (2). An ideal method for EBL and EHL should be quick, easy and accurate.
Currently, AI technology is increasingly applied in the medical field. Recently, Triton System, which is based on AI, has been introduced to several hospitals in the USA. This device is able to dynamically estimate intra-operative blood loss and Hb loss. Referring to the patent (8), the actual Hb mass in images used for model construction was detected by rinse methods; while the actual blood loss calculated by Triton System was used to estimate the Hb mass and available Hb concentration of the patient, which are not affected by other fluids. The EBL by Triton System was reported to have a strong correlation with photometric methods (3,4,6). However, the equation used to calculate the EBL presented certain limits; for example, the Hb concentration of the patient could change via fluid therapy or blood loss. Unfortunately, the research involving Triton System did not present indicators for model performance, such as MAE, MSE and R-squared. However, the root mean squared error (RMSE), which indicates the measurement error of the model, of Triton System was 1.15 g per sponge, which was larger than that of ours (
In the current study, the methods based on Xgboost and DenseNet achieved the best performance regardless of being used for EBL or EHL. Xgboost is a well-known modified algorithm that can construct an optimal model by minimizing the loss function. However, Xgboost possessed worse 95% CIs for model performance and concordance analysis results than those of DenseNet, which may be due to the less satisfactory model generalization ability of Xgboost. Compared with feature engineering, apart from the advantages of traditional deep learning methods, DenseNet has the following main advantages (11): to achieve the same accuracy, DenseNet requires fewer parameters. DenseNet has a very good resistance to over-fitting, especially for applications where training data are relatively scarce and its generalization performance is stronger, so if there is no data augmentation, DenseNet does not drop significant data. Therefore, despite the number of images in our study not being large enough for typical deep learning methods, DenseNet achieved a satisfactory performance. The results indicated that the research and development costs would be lower than those based on traditional feature engineering. It is strongly suggested that in the near future, our methods would be substantially cheaper to promote.
In the current study, we attempted to develop a better method based on AI to estimate intra-operative blood loss, not to validate a new device or software. Therefore, there are some limitations. First, we only applied AI technology for EBL and EHL on one kind of sponge. Research on EBL and EHL on other sizes of sponges and canisters should be performed. Second, due to the difficulties of collecting non-recycled blood, we only set the volume gradient from 1 to 6 mL. However, for AI technology, the more images there are, the better model; nevertheless, based on the current images, relatively satisfied results were achieved. In the near future, we will introduce more images to elevate the model performance.
Conclusions
In the current study, we developed a new method for estimating blood loss and Hb loss based on DenseNet, which achieved a higher accuracy and lower bias than those of methods based on feature engineering. The presented model may be a better AI-based method for the estimation of intra-operative blood and Hb loss, especially for surgeries primarily using sponges and for a small to medium amount of blood loss.
Supplementary
Establishment of blood-soaked sponges
In the current study, we set the gradient of blood as following: 1, 2, 3, 4, 5, 6 mL. The typical view of the blood-soaked sponges were shown in Figure S1.
Image pre-processing
To estimate intraoperative blood loss on sponge, the first factor to be considered is the area of the blood. In the proposed method, the area of blood is divided into two parts for extraction. Firstly, the blood-soaked sponge image is reshaped to 480×480×3 and converted from RGB color space to HSV color space. The value of pixel in H channel ranges from 0 to 180. The value of pixel in S channel and V channel ranges from 0 to 255. Secondly, two mask maps are generated by Eq. [2] and Eq. [3], where the M1ij and M2ij are the pixels at position (i, j) of two mask maps, the Hij, Sij and Vij are the pixels at position (i, j) of H, S and V channels.
[2]
[3]
Then, the two mask maps are used to generate the two blood areas by Eq. [4] and Eq. [5], where B ij is the pixel vector at position (i, j) of the blood-soaked sponge image, BR1 ij and BR2 ij are the pixel vector at position (i, j) of the two blood areas. Figure S2 shows the tow blood areas images and the blood-soaked sponge image, it is obviously that the two blood area images only have two colors red and black, and the red area is represented the blood area.
[4]
[5]
Feature extraction
After get the two blood area images B1 and B2, the two area features and other color features can be calculated. The first step is to convert the two blood area images from RGB color space to HSV color space. Since the two blood area images only have two colors red and black. As a result, the two area features can be represented by the red area in the image to the overall image area. In order to simplify the extraction process, the ratio of the number of non-0 pixels in the H plane to the number of total pixels was used to approximate the area ratio. Assuming the number of the non-0 pixels in the two blood areas is RP1num and RP2num, then the two area features can be calculated by Eq. [6].
[6]
In addition to area, the color depth and brightness of the blood area are also important factors in estimating intraoperative blood loss on gauze. In HSV color space, the three channels represent hue, saturation and brightness respectively. Therefore, the proposed method uses the mean and variance of H, S and V channels to represent the color depth and brightness features of the blood area. These features can be calculated by Eq. [7], where m and n was the size of image, hi,j, si,j and vi,j represent the pixel of H, S and V channels in position (i, j) respectively. Finally, the features of the blood-soaked gauze image have fourteen elements and were shown in Table S1. Tables S2 and S3 show the hyperparameters of random forest and Xgboost used in feature engineering method.
[7]
Supercomputing platform
The experimental environment of this article was based on the GPU supercomputing cluster server: consists of three FitServer R4200, Ubuntu 16.04 LTS was used as the operating system with Intel Xeon e5-2620 V4 processor and Nvidia GTX 1080 Ti GPU, the memory is 128 GB. Pytoch was used to build the convolutional neural network, and Python3.6 was used as the programming language (shown in Figure S3).
Acknowledgments
Funding: This work was supported by National Key R&D Program of China (No. 2018YFC0116702 and No. 2018YFC0116704), National Natural Science Foundation of China (No. 81870422 and No. 81600035), Medical Innovation Capacity Improvement Program for Medical Staff of the First Affiliated Hospital of the Third Military Medical University (No. SWH2018QNKJ-27), Technology Innovation and Application Research and Development Project of Chongqing City (cstc2019jscx-msxmX0237).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/atm-20-1806
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-1806
Peer Review File: Available at http://dx.doi.org/10.21037/atm-20-1806
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-1806). Dr. YJL, LGZ, HYZ, KHZ, ZYY, KZL, JZ, YWC, BY have a patent 202010324328.5 pending. The other authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study protocol was approved by the Institutional Ethics Committee of Southwest Hospital of Third Military Medical University (No. KY2019127) on October 28, 2019. Written informed consent was obtained from each patient.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Solon JG, Egan C, McNamara DA. Safe surgery: how accurate are we at predicting intra-operative blood loss? J Eval Clin Pract 2013;19:100-5. [Crossref] [PubMed]
- Schorn MN. Measurement of blood loss: review of the literature. J Midwifery Womens Health 2010;55:20-7. [Crossref] [PubMed]
- Nowicki PD, Ndika A, Kemppainen J, et al. Measurement of Intraoperative Blood Loss in Pediatric Orthopaedic Patients: Evaluation of a New Method. J Am Acad Orthop Surg Glob Res Rev 2018;2:e014. [Crossref] [PubMed]
- Konig G, Waters JH, Javidroozi M, et al. Real-time evaluation of an image analysis system for monitoring surgical hemoglobin loss. J Clin Monit Comput 2018;32:303-10. [Crossref] [PubMed]
- Konig G, Waters JH, Hsieh E, et al. In Vitro Evaluation of a Novel Image Processing Device to Estimate Surgical Blood Loss in Suction Canisters. Anesth Analg 2018;126:621-8. [Crossref] [PubMed]
- Holmes AA, Konig G, Ting V, et al. Clinical evaluation of a novel system for monitoring surgical hemoglobin loss. Anesth Analg 2014;119:588-94. [Crossref] [PubMed]
- Adkins AR, Lee D, Woody DJ, et al. Accuracy of blood loss estimations among anesthesia providers. AANA J 2014;82:300-6. [PubMed]
- Siddarth Satish S, Miller K, Zandifar A. System and Method for estimating extracorporeal blood volume in a physical sample. USA, 2013.
- Saggu M, Patel AR, Koulis T. A Random Forest Approach for Counting Silicone Oil Droplets and Protein Particles in Antibody Formulations Using Flow Microscopy. Pharm Res 2017;34:479-91. [Crossref] [PubMed]
- Tahmassebi A, Wengert GJ, Helbich TH, et al. Impact of Machine Learning With Multiparametric Magnetic Resonance Imaging of the Breast for Early Prediction of Response to Neoadjuvant Chemotherapy and Survival Outcomes in Breast Cancer Patients. Invest Radiol 2019;54:110-7. [Crossref] [PubMed]
- Huang G, Liu Z, van der Maaten L. Densely Connected Convolutional Networks. 2016.
- Tong N, Gou S, Yang S, et al. Shape constrained fully convolutional DenseNet with adversarial training for multiorgan segmentation on head and neck CT and low-field MR images. Med Phys 2019;46:2669-82. [Crossref] [PubMed]
- Škrabánek P, Zahradníková A Jr. Automatic assessment of the cardiomyocyte development stages from confocal microscopy images using deep convolutional networks. PLoS One 2019;14:e0216720. [Crossref] [PubMed]
- Chen TG, Carlos. XGBoost: A Scalable Tree Boosting System. 2016.
- Ashburn JC, Harrison T, Ham JJ, et al. Emergency physician estimation of blood loss. West J Emerg Med 2012;13:376-9. [Crossref] [PubMed]
- Rothermel LD, Lipman JM. Estimation of blood loss is inaccurate and unreliable. Surgery 2016;160:946-53. [Crossref] [PubMed]
- Duthie SJ, Ven D, Yung GL, et al. Discrepancy between laboratory determination and visual estimation of blood loss during normal delivery. Eur J Obstet Gynecol Reprod Biol 1991;38:119-24. [Crossref] [PubMed]
- Bose P, Regan F, Paterson-Brown S. Improving the accuracy of estimated blood loss at obstetric haemorrhage using clinical reconstructions. BJOG 2006;113:919-24. [Crossref] [PubMed]
- Ali Algadiem E, Aleisa AA, Alsubaie HI, et al. Blood Loss Estimation Using Gauze Visual Analogue. Trauma Mon 2016;21:e34131. [Crossref] [PubMed]
- Zuckerwise LC, Pettker CM, Illuzzi J, et al. Use of a novel visual aid to improve estimation of obstetric blood loss. Obstet Gynecol 2014;123:982-6. [Crossref] [PubMed]