Novel mutation of GJA8 in autosomal dominant congenital cataracts
Introduction
Congenital cataract, which is a significant cause of childhood blindness, seriously affects the development of children’s vision and can lead to blindness or amblyopia in infants. It is estimated that the incidence rate is between 1 and 6 per 10,000 infants. Worldwide, congenital cataracts account for the rate of blindness in children is 5–20% in developed countries and 22–30% in developing countries (1).
The causes of congenital cataract are complex, and includes genetics, pregnancy infections, metabolic diseases, and chromosomal abnormalities, with some still being unknown. Congenital cataract can occur as an isolated ocular disease or as a part of genetic syndromes accompanied by multisystemic abnormalities. In another study, of 243 children with newly diagnosed congenital or infantile cataract, hereditary disease was associated with 56% of bilateral cases but only 6% of unilateral cases (2). Approximately 50% of all congenital cataract cases may have a genetic cause, and such cases are quite heterogeneous. Genetic mutation, especially autosomal dominant transmission, remains the most common cause. A number of protein-coding genes have been reported including crystallins, lens-specific connexins, aquaporin, cytoskeletal structural proteins, and developmental regulators (3). More than 100 genes have been identified in congenital cataracts, which can be divided into syndromic and nonsyndromic cataracts genes (4). About half of the mutations occurred in the crystallin genes, and a quarter in membrane proteins genes (5), such as connexins (Cxs) and major intrinsic proteins (MIPs). Mutations in Cxs account for around 16% of cases with a known genetic cause (6). Connexin43, connexin46, and connexin50 are abundant in human lens. Connexin43 mainly expressed in lens epithelial cells (7). Cx46, encoded by GJA3, is expressed only in fiber cells, whereas Cx50, encoded by GJA8, is present throughout the lens (8). Any significant dysregulation of GJA8 could be deleterious for eye development.
Congenital cataract appears at birth or during early childhood. Visual deprivation caused by congenital cataracts can damage the development of visual function. However, there are no effective measures clinically to prevent the onset of the disease, and currently, the main treatment is cataract removal. Therefore, early detection and prompt surgical intervention of congenital cataracts is crucial to prevent irreversible amblyopia and promote visual reconstruction (9). With the progress of microincision surgical technique, cataract removal combined with anterior vitrectomy and primary intraocular lens (IOL) implantation has been approved as the main surgical method for congenital cataracts (10). However, some postoperative complications may occur after surgery in children, such as inflammatory response, secondary glaucoma and posterior capsule opacification (PCO), which still affect the development of visual function if poorly controlled. Moreover, primary IOL implantation is recommended for children over 2 years old, but it is a challenge for ophthalmologists to predict the development of infant eyeballs and choose the appropriate diopter of IOL, as unstable axial length, changeable corneal curvature, myopia drift and other factors should be considered comprehensively (11-13). Accordingly, Genetic screening during pregnancy is of great significance for the early diagnosis and prevention of congenital cataract.
In this study, we sought to identify the genetic defect of autosomal dominant congenital cataracts (ADCC) in a Chinese family. Using high-throughput sequencing technology, whole exome sequencing and pathogenic gene analysis were performed. A novel missense mutation was identified in this family. This data expands the spectrum of GJA8 variants likely to be contributing to eye anomalies and the spectrum of phenotypes associated with this gene. We then identified the correlation between the pathogenic genes and the disease, so as to provide a theoretical basis for further clinical prenatal diagnosis, early intervention or gene therapy in the future. In addition, it remains to be studied whether this site is a hotspot or high frequency variant of multiple cataract type 1 caused by GJA8 gene mutation in Chinese population. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/atm-20-4663).
Methods
Patients
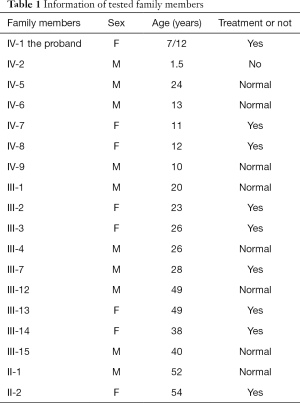
This study was approved by the Ethics Committee of Beijing Tongren Hospital, Capital Medical University. It was performed in accordance to the tenets of the Declaration of Helsinki (as revised in 2013). The recruited family comprised 34 affected individuals from a four-generation pedigree, 33 of whom were present, including 17 patients, 5 males, and 12 females (Figure 1). According to the history and medical records, all affected members except the first generation were diagnosed before the age of 2, and received cataract surgery before the age of 5. They had different degrees of strabismus amblyopia, while one of them occurred secondary glaucoma. The proband’s mother denied any history of inbreeding, infection, medication during pregnancy, or preterm birth. Blood samples from 10 patients and 8 normal relatives were collected from the proband and her available family members after informed consent was obtained (Table 1). Clinical examinations were performed by ophthalmologists.
Full table
Study methods
DNA extraction
For DNA extraction, 5-mL samples of blood were collected into vacuum tubes containing an anticoagulant agent (EDTA) and stored at −4 °C. Genomic DNA of proband and family members was extracted using Universal Gen DNA Kit for quality testing.
Targeted capture and high-throughput sequencing
The genome DNA of the proband was sheared into fragments using a Covaris crusher. A DNA library was constructed, after which whole exome sequencing was performed. We used the high-throughput targeted capture for further bioinformatics analysis to determine the genes related to hereditary eye diseases.
Mapping reads and annotating variant positions
The clean data was obtained by adapter trimming and filtering the low-quality reads from the raw data. Using a comparison to the reference genome with the Burrows-Wheeler aligner (BWA), we evaluated the quality of sequencing data, and then analyzed the mutation results. The annotated data were combined with clinical phenotypes analysis, and, the variations were further screened and interpreted in detail.
The naming rules were based on the Human Genome Variation Society (HGVS), and the interpretation and evidence grading of variation were performed according to the latest edition of the standards and guidelines for the interpretation of gene variation issued by the American Society for Medical Genetics and Genomics (ACMG) (14).
The analysis results of clean data showed that the coverage of the target area was 99.69%, the average sequencing depth was 119.1×, and the coverage of the sequencing depth greater than 30× was 96.64%. It indicated that the target area had been effectively covered.
Sanger sequencing
The c.178G>A site in gap junction protein alpha 8 (GJA8) gene of 18 family members was amplified by polymerase chain reaction (PCR), and DNA was detected by using the ABI 3730xL DNA genetic analyzer. Finally, the sequencing results were recorded in Chromas and DNA Star software for sequence comparison. The reference sequence NG_016242.1 was obtained from National Center of Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/nuccore/NG_016242.1?report=genbank&from=4976&to=11451).
In this study, the primers of GJA8 gene were designed by software Oligo and Primer-BLAST and synthesized by Sangon Biotech (Shanghai) Co., Ltd. (Table 2).
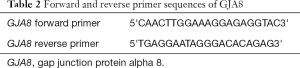
Full table
Results
Clinical manifestations
Affected family members were found in each generation in the recruited family, with the prevalence ratio of males to females being 1:2.5. Pedigree analysis suggested an autosomal-dominant inheritance pattern in this family (Figure 1).
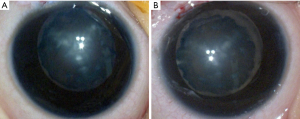
The proband is a 7-month-old female (IV-1, Figure 1). She suffered from a bilateral symmetric opacity of the lens at birth, with Y-shaped nuclear opacification, but this did not progress with age. Her right eye is more obviously afflicted than her left eye (Figure 2). There was no family history of other ocular or systemic abnormalities. Other family members have similar cataract patterns that were either received at birth or in the earlier years after birth.
Whole exome sequencing of the proband
After a comparison with the genetic variation sites in dbSNP, The 1000 Genomes Project, ExAC, OMIM, HGMD, ClinVar, and other East Asian population databases, variation sites which frequency is not higher than 0.01 were selected, the genes related to hereditary eye diseases were analyzed, and the following candidate mutation gene loci were screened out: GJA8 gene c.178G>A; ACO2 gene c.1387G>T; CYP4V2 gene c.1354C>T; HMCN1 gene c.5296A>G; NEK2 gene c.76C>T; PRDM5 gene c.269C>T; RGR gene c.744+5A>G; USH2A gene c.8306C>T.
All novel findings were evaluated according to ACMG standards and guidelines for further screening and detailed interpretation of mutation sites combined with clinical phenotype and family verification. Candidate variations that were significantly inconsistent with the clinical phenotype were eliminated and whether they conform to the genetic law were determined. The results strongly suggested that GJA8 gene c.178G>A was a possibly pathogenic mutation, and this accorded with the patient clinical phenotype and genetic pattern of the family.
Sanger sequencing results
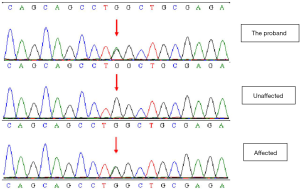
Sanger sequencing verified the site of GJA8 gene c.178G>A of 18 family members, and found that, in all patients with the cataract phenotype in this family, the 178th nucleotide of GJA8 gene changed from G to A, and the mutation site and type were the same as that of the proband; meanwhile, this gene mutation was not found in the family members with a normal phenotype (Figure 3).
Discussion
Congenital cataract is one of the significant causes of child blindness and visual impairment, and shows obvious genetic heterogeneity. Approximately 70% of familial, sporadic or non-syndrome cases can obtain genetic diagnoses using genomics methods (15).
GJA8 is a lens gap junction protein whose function includes moderating the diffusion of molecules between the lens fiber cells. GJA8 mutations are known to cause microphthalmia, microphakia and congenital cataracts. More than 30 mutations have been reported (16), most of which are nuclear cataracts. Gap junctional complexes are also present in the epithelium, fibroblasts, and endothelium of the cornea. Other studies have found that GJA8 mutations can also lead to severe congenital corneal opacifications, sclerocornea, or lens abnormalities including congenital cataracts (17).
Currently, mutations in a number of genes have been reported to cause congenital cataract. In this study, we recruited an ADCC family. Gene mutations were analyzed by targeted capture and high-throughput sequencing, and were used to compare the unaffected and affected family members. Mutations that were meaningless or that caused other diseases were excluded. Suspected mutation sites of the proband were screened in the target genes. We found that GJA8 gene c.178G>A is the pathogenic mutation of the pedigree.
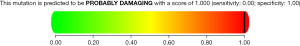
A heterozygous missense mutation in GJA8 gene c.178G>A was identified in the proband and confirmed in the affected family members. Gene sequencing revealed the missense mutation in exon 2 of the GJA8 gene, from glycine to serine at position 178 (p.G60S). No significant mutation was found in other genes related to congenital cataract. The mutation was detected within the hotspot. It was not found in the ExAC browser or the 1,000 Genomes Project databases of the East Asian population. Furthermore, the mutation position was highly conserved. Bioinformatics software was used to analyze the possible influence on protein structure and function, and all results showed that the novel mutation might be pathogenic. The mutation was predicted to be disease causing by Mutation Taster. The Polyphen2 score was 1.00 (Figure 4), which suggests that this mutation is probably damaging and is likely to affect the function of the protein. The SIFT score was 0.00, which also indicates that the mutation is damaging and affects protein function. Multiple cataract type 1 caused by GJA8 gene mutation is autosomal dominant inheritance. The clinical phenotype of the disease is consistent with that of the proband. The family investigation showed that co-segregation of the disease phenotype and the detected variants were verified in all family members. This heterozygous mutation was detected in all patients in the family, which confirmed by Sanger sequencing, but not in ones with the normal phenotype. According to the standards and guidelines for the interpretation of gene variation issued by the ACMG, the variation can be considered to be a possibly pathogenic mutation.
In conclusion, we identified a pathogenic mutation of c.178G>A in the GJA8 gene of an ADCC family. These findings further expand the mutation spectrum of the GJA8 gene, verify the important role of GJA8 gene in lens development, and provide useful information for congenital cataract in molecular biology. Moreover, our results make it possible to explore whether this mutation leads to a change of the protein structure and function in the future studies. This information may not only be relevant for guiding the accurate genetic counseling, the individualized diagnosis of congenital cataracts and prenatal screening in affected family members to have a second child, but also can provide a reference for the mutation gene screening for other congenital cataract families.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/atm-20-4663
Data Sharing Statement: Available at http://dx.doi.org/10.21037/atm-20-4663
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-4663). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Ethics Committee of Beijing Tongren Hospital, Capital Medical University (TREC2009-39). It was performed in accordance to the tenets of the Declaration of Helsinki (as revised in 2013) and informed consent was obtained from all participants.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Liu YC, Wilkins M, Kim T, et al. Cataracts. Lancet 2017;390:600-12. [Crossref] [PubMed]
- Rahi JS, Dezateux C. Congenital and infantile cataract in the United Kingdom: underlying or associated factors. British Congenital Cataract Interest Group. Invest Ophthalmol Vis Sci 2000;41:2108-14. [PubMed]
- Pichi F, Lembo A, Serafino M, et al. Genetics of Congenital Cataract. Dev Ophthalmol 2016;57:1-14. [Crossref] [PubMed]
- Messina-Baas O, Cuevas-Covarrubias SA. Inherited Congenital Cataract: A Guide to Suspect the Genetic Etiology in the Cataract Genesis. Mol Syndromol 2017;8:58-78. [Crossref] [PubMed]
- Bari KJ, Sharma S, Chary KVR. Structure of G57W mutant of human γS-crystallin and its involvement in cataract formation. J Struct Biol 2019;205:72-8. [Crossref] [PubMed]
- Shiels A, Hejtmancik JF. Mutations and mechanisms in congenital and age-related cataracts. Exp Eye Res 2017;156:95-102. [Crossref] [PubMed]
- Jiang JX. Gap junctions or hemichannel-dependent and independent roles of connexins in cataractogenesis and lens development. Curr Mol Med 2010;10:851-63. [Crossref] [PubMed]
- Ceroni F, Aguilera-Garcia D, Chassaing N, et al. New GJA8 variants and phenotypes highlight its critical role in a broad spectrum of eye anomalies. Hum Genet 2019;138:1027-42. [Crossref] [PubMed]
- Li J, Chen X, Yan Y, et al. Molecular genetics of congenital cataracts. Exp Eye Res 2020;191:107872. [Crossref] [PubMed]
- Solebo AL, Hammond CJ, Rahi JS. Improving outcomes in congenital cataract. Nature 2018;556:E1-2. [Crossref] [PubMed]
- Lambert SR. Changes in Ocular Growth after Pediatric Cataract Surgery. Dev Ophthalmol 2016;57:29-39. [Crossref] [PubMed]
- Seven E, Tekin S, Batur M, et al. Evaluation of changes in axial length after congenital cataract surgery. J Cataract Refract Surg 2019;45:470-4. [Crossref] [PubMed]
- Valera Cornejo DA, Flores Boza A. Relationship between preoperative axial length and myopic shift over 3 years after congenital cataract surgery with primary intraocular lens implantation at the National Institute of Ophthalmology of Peru, 2007-2011. Clin Ophthalmol 2018;12:395-9. [Crossref] [PubMed]
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. [Crossref] [PubMed]
- Ma AS, Grigg JR, Ho G, et al. Sporadic and Familial Congenital Cataracts: Mutational Spectrum and New Diagnoses Using Next-Generation Sequencing. Hum Mutat 2016;37:371-84. [Crossref] [PubMed]
- Beyer EC, Ebihara L, Berthoud VM. Connexin mutants and cataracts. Front Pharmacol 2013;4:43. [Crossref] [PubMed]
- Ma AS, Grigg JR, Prokudin I, et al. New mutations in GJA8 expand the phenotype to include total sclerocornea. Clin Genet 2018;93:155-9. [Crossref] [PubMed]